Bioorganic & Medicinal Chemistry ( IF 3.3 ) Pub Date : 2021-02-18 , DOI: 10.1016/j.bmc.2021.116070 Tomoko Furuzono 1 , Asako Murata 2 , Satoshi Okuda 3 , Kenji Mizutani 3 , Tsuyoshi Adachi 3 , Kazuhiko Nakatani 2
|
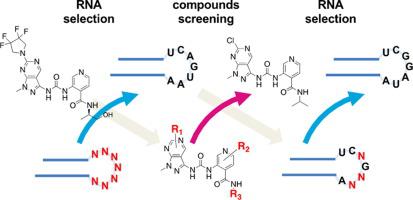
RNA is an emerging target of next-generation drug development. Recently, new small molecules targeting RNAs were discovered by several pharmaceutical companies. Methods have been reported to identify small molecules targeting a specific RNA sequence and structural motif, however, because of diverse sequence and structural motifs potentially present in the druggable functional RNAs, large sets of structure-activity relationships (SARs) information of small molecule – RNA interactions will be required for the acceleration and efficient startup of the discovery programs toward unprecedented RNA targets. Here we describe our iterative RNA selection and compounds screening to accumulate rich information about small molecules – RNA interaction. The RNAs that selectively bind to the initial molecular target, compound 1 from our in-house chemical library (JT-library), was isolated using in vitro selection technique from a hairpin-structured RNA library mimicking precursor-microRNA (pre-miRNA). Then, we engineered pre-let-7f-2 to create its mutant that can bind to compound 1 by embedding the in vitro selected RNA motif for compound 1 in the hairpin loop region. The obtained mutant pre-let-7f-2-loop-mt was used as a target for screening 316 analogs of compound 1. A surface plasmon resonance (SPR) -based screening was performed against pre-let-7f-2-loop-mt-immobilized sensor surface and we obtained four compounds that can bind to the RNA. Among these four compounds, three compounds showed higher affinity to pre-let-7f-2-loop-mt than the parental compound 1, which suggests the feasibility of our strategy for gathering the SAR information on small molecule – RNA interactions. We demonstrated only one cycle of RNA selection and compound screening in the present study, but can continue this cycle with the selected molecule to gain new RNAs and even new RNA motifs and gather much SAR information with improved accuracy.
中文翻译:

加速靶向 RNA 的药物发现:用于探索 RNA-小分子对的迭代“RNA 选择-化合物筛选循环”
RNA是下一代药物开发的新兴靶点。最近,几家制药公司发现了新的靶向 RNA 的小分子。已经报道了鉴定靶向特定 RNA 序列和结构基序的小分子的方法,但是,由于可成药的功能性 RNA 中可能存在不同的序列和结构基序,小分子 - RNA 的大量构效关系 (SAR) 信息加速和有效启动针对前所未有的 RNA 目标的发现程序需要相互作用。在这里,我们描述了我们的迭代 RNA 选择和化合物筛选,以积累关于小分子 - RNA 相互作用的丰富信息。选择性结合初始分子靶标化合物1的 RNA从我们的内部化学文库(JT 文库)中分离,使用体外选择技术从模拟前体微 RNA(pre-miRNA)的发夹结构 RNA 文库中分离。然后,我们工程改造的预让-7F-2以创建其突变体能够结合到化合物1通过嵌入化合物的体外筛选的RNA基序1的发夹环区。获得的突变体pre-let-7f-2-loop-mt被用作筛选化合物1的316个类似物的靶标。针对 pre-let-7f-2-loop-mt 固定化传感器表面进行基于表面等离子体共振 (SPR) 的筛选,我们获得了四种可以与 RNA 结合的化合物。在这四种化合物中,三种化合物对 pre-let-7f-2-loop-mt 的亲和力高于母体化合物1,这表明我们收集关于小分子 - RNA 相互作用的 SAR 信息的策略的可行性。我们在本研究中仅展示了一个 RNA 选择和化合物筛选循环,但可以用选定的分子继续这个循环,以获得新的 RNA 甚至新的 RNA 基序,并以更高的准确性收集大量 SAR 信息。











































 京公网安备 11010802027423号
京公网安备 11010802027423号