Talanta Open ( IF 4.1 ) Pub Date : 2021-02-12 , DOI: 10.1016/j.talo.2021.100033 Jiayu Wang , Xuetong Bi , Wei Chen , Qinyue Zhao , Jinqi Yang , Xiangjun Tong , Meiping Zhao
|
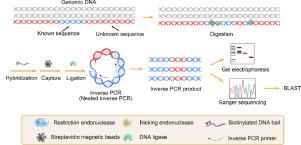
We demonstrate a novel method for identification of the insertion site of transgenic DNA by construction a circular DNA containing part of the transgenic DNA (tDNA) and its flanking sequence, which would be different from those in the native genomic DNA (gDNA). Using follistatin (FST) as a model of tDNA, we cleaved the sequence of FST gene and the adjacent sequence with a proper combination of restriction endonuclease and nicking endonuclease and isolated the resultant fragments containing the target FST gene from the gDNA background. Two types of previously unknown ligation property of T7 DNA ligase were disclosed in our experiments. One is non-templated single-stranded overhang ligation capability; the other one is the successful ligation of 3’-overhang and 5’-recessed ends in the presence of two mismatched base pairs. By virtue of these efficient ligation reactions, we successfully construct the circular DNA with high cyclization yield. Further coupling with nested inverse PCR, the method enabled identification of transgenic follistatin a (fsta) cDNA in zebrafish embryonic DNA. The established method provides a practical tool to determine the insertion site of tDNA with a sensitivity of 103 copies in 1.0 μg of gDNA and high specificity. It may be further extended for gene doping detection and assessment of the safety of gene therapy and genetically modified organisms.
中文翻译:

基于带有侧翼序列的靶基因环化和巢式反向PCR,鉴定转基因DNA的插入位点
我们展示了一种通过构建包含部分转基因DNA(tDNA)及其侧翼序列的环状DNA来鉴定转基因DNA插入位点的新颖方法,这将不同于天然基因组DNA(gDNA)中的那些。使用卵泡抑素(FST)作为tDNA的模型,我们用限制性内切核酸酶和切口内切核酸酶的适当组合切割了FST基因的序列和相邻序列,并分离出包含目标FST的片段基因来自gDNA背景。在我们的实验中公开了T7 DNA连接酶的两种先前未知的连接特性。一种是非模板化的单链突出端连接能力;另一个是在存在两个错配碱基对的情况下成功连接3'突出端和5'凹陷端。通过这些有效的连接反应,我们成功地构建了具有高环化产率的环状DNA。进一步与嵌套反向PCR结合,该方法能够鉴定斑马鱼胚胎DNA中的转基因卵泡抑素α(fsta)cDNA。所建立的方法为确定tDNA的插入位点提供了一种实用的工具,其灵敏度为10 3可在1.0μggDNA中复制且具有高特异性。它可能会进一步扩展到基因掺杂检测以及基因治疗和转基因生物的安全性评估。











































 京公网安备 11010802027423号
京公网安备 11010802027423号