Current Bioinformatics ( IF 4 ) Pub Date : 2020-11-30 , DOI: 10.2174/1574893615999200429121156 Kanu Geete 1 , Manish Pandey 1
|
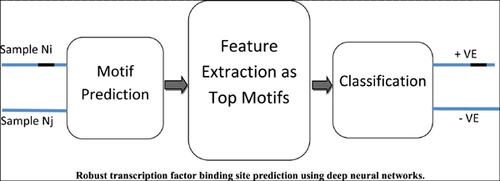
Aim: Robust and more accurate method for identifying transcription factor binding sites (TFBS) for gene expression.
Background: Deep neural networks (DNNs) have shown promising growth in solving complex machine learning problems. Conventional techniques are comfortably replaced by DNNs in computer vision, signal processing, healthcare, and genomics. Understanding DNA sequences is always a crucial task in healthcare and regulatory genomics. For DNA motif prediction, choosing the right dataset with a sufficient number of input sequences is crucial in order to design an effective model.
Objective: Designing a new algorithm which works on different dataset while an improved performance for TFBS prediction.
Methods: With the help of Layerwise Relevance Propagation, the proposed algorithm identifies the invariant features with adaptive noise patterns.
Results: The performance is compared by calculating various metrics on standard as well as recent methods and significant improvement is noted.
Conclusion: By identifying the invariant and robust features in the DNA sequences, the classification performance can be increased.
中文翻译:

使用深度神经网络的鲁棒转录因子结合位点预测
目的:一种可靠且更准确的方法,用于识别基因表达的转录因子结合位点(TFBS)。
背景:深度神经网络(DNN)在解决复杂的机器学习问题方面显示出令人鼓舞的增长。在计算机视觉,信号处理,医疗保健和基因组学领域,DNN轻松地取代了传统技术。了解DNA序列始终是医疗保健和监管基因组学中的关键任务。对于DNA基序预测,选择正确的具有足够数量输入序列的数据集对于设计有效模型至关重要。
目的:设计一种适用于不同数据集的新算法,同时提高TFBS预测的性能。
方法:在分层相关传播的帮助下,提出的算法利用自适应噪声模式识别不变特征。
结果:通过计算各种标准和最新方法的指标来比较性能,并指出了显着改进。
结论:通过鉴定DNA序列的不变和鲁棒的特征,可以提高分类性能。



























 京公网安备 11010802027423号
京公网安备 11010802027423号