Current Bioinformatics ( IF 2.4 ) Pub Date : 2020-11-30 , DOI: 10.2174/1574893615999200626190346 Wenwen Ran 1 , Xiangbin Chen 2 , Bo Wang 2 , Ping Yang 3 , Yongxing Li 1 , Yujing Xiao 1 , Xiaonan Wang 1 , Guangqi Li 1 , Lili Wang 1 , Yingmin Han 2 , Yonggang Peng 2 , Jidong Lang 2 , Yuebin Liang 2 , Yupei Xiao 2 , Qingqing Lu 2 , Huixin Lin 2 , Geng Tian 2 , Dawei Yuan 2 , Chaoyang Deng 2 , Jialiang Yang 2 , Xiaoming Xing 1
|
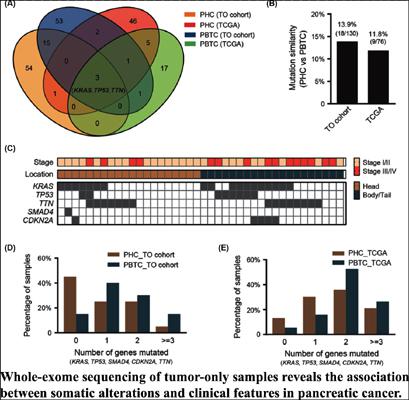
Background: Identification of genomic markers using NGS (next-generation sequencing) technology would be valuable for guiding precision medicine treatments for pancreatic cancers. Traditional somatic mutation methods require both tumor and matched non-tumor samples. However, only tumor samples are available mostly, especially in retrospective studies. In this study, we tried to analyze the associations between clinical features and oncogenic somatic mutations in genome-wide tumor-only samples.
Methods: Fifty-four tumor-only samples derived from pancreatic cancer patients were used for whole-exome sequencing. An approach involving SNP filtering of variants included in the Catalogue of Somatic Mutations in Cancer (COSMIC) database was used to identify oncogenic somatic mutations. The relationships between oncogenic mutations and clinical features were analyzed and simultaneously compared with those from the TCGA database.
Results: By analyzing the mutations from tumor only samples, divergent mutation profiles were observed in different locations (head vs. body/tail) of pancreatic tumors. The divergences between pancreatic head and body/tail cancers were also confirmed by the TCGA data. Furthermore, mutations of several genes were found to be significantly associated with clinical features, such as pathological stage and the degree of tumor differentiation.
Conclusion: The results confirmed the efficiency of our approach in identifying oncogenic somatic mutations from tumor only samples and revealed the associations between somatic mutations and clinical features in pancreatic cancer.
中文翻译:

仅肿瘤样本的全外显子测序揭示了胰腺癌的体细胞变异与临床特征之间的关联
背景:使用NGS(下一代测序)技术鉴定基因组标记物对于指导胰腺癌的精确药物治疗将具有重要价值。传统的体细胞突变方法需要肿瘤和匹配的非肿瘤样品。但是,大多数情况下仅可获得肿瘤样品,尤其是在回顾性研究中。在这项研究中,我们试图分析全基因组仅肿瘤样本中临床特征与致癌体细胞突变之间的关联。
方法:将来自胰腺癌患者的54个仅肿瘤样本用于全外显子组测序。癌症体细胞突变目录(COSMIC)数据库中包含的涉及变异体SNP过滤的方法用于鉴定致癌体细胞突变。分析了致癌突变与临床特征之间的关系,并与TCGA数据库进行了比较。
结果:通过分析仅肿瘤样品的突变,在胰腺肿瘤的不同位置(头部与身体/尾巴)观察到了不同的突变谱。TCGA数据也证实了胰头癌和身体/尾巴癌之间的差异。此外,发现几种基因的突变与临床特征显着相关,例如病理阶段和肿瘤分化程度。
结论:结果证实了我们的方法从仅肿瘤样品中鉴定致癌性体细胞突变的有效性,并揭示了体细胞突变与胰腺癌临床特征之间的关联。











































 京公网安备 11010802027423号
京公网安备 11010802027423号