Current Bioinformatics ( IF 4 ) Pub Date : 2020-11-30 , DOI: 10.2174/1574893615999200706002907 Yeqing Sun 1 , Lei Chen 1 , Yingqi Zhang 1 , Jincheng Zhang 1 , Shashi Ranjan Tiwari 1
|
Background: Osteoarthritis (OA), one of the most important causes leading to joint disability, was considered as an untreatable disease. A series of genes were reported to regulate the pathogenesis of OA, including microRNAs, Long non-coding RNAs and Circular RNA. So far, the expression profiles and functions of lncRNAs, mRNAs, and circRNAs in OA are not fully understood.
Objective: The present study aimed to identify differentially expressed genes in OA.
Methods: The present study conducted RNA-seq to identify differentially expressed genes in OA. Ontology (GO) analysis was used to analyze the Molecular Function and Biological Process. KEGG pathway analysis was used to perform the differentially expressed lncRNAs in biological pathways.
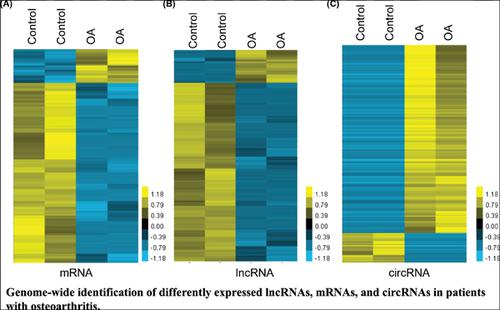
Results: Hierarchical clustering revealed a total of 943 mRNAs, 518 lncRNAs, and 300 circRNAs, which were dysregulated in OA compared to normal samples. Furthermore, we constructed differentially expressed mRNAs mediated protein-protein interaction network, differentially expressed lncRNAs mediated trans-regulatory networks, and competitive endogenous RNA (ceRNA) to reveal the interaction among these genes in OA. Bioinformatics analysis revealed that these dysregulated genes were involved in regulating multiple biological processes, such as wound healing, negative regulation of ossification, sister chromatid cohesion, positive regulation of interleukin-1 alpha production, sodium ion transmembrane transport, positive regulation of cell migration, and negative regulation of inflammatory response. To the best of our knowledge, this study for the first time, revealed the expression pattern of mRNAs, lncRNAs and circRNAs in OA.
Conclusion: This study provided novel information to validate these differentially expressed RNAs may be as possible biomarkers and targets in OA.
中文翻译:

骨关节炎患者中全基因组差异表达的lncRNA,mRNA和circRNA的鉴定
背景:骨关节炎(OA)是导致关节残疾的最重要原因之一,被认为是一种无法治愈的疾病。据报道,一系列基因可调节OA的发病机制,包括microRNA,长非编码RNA和环形RNA。到目前为止,尚未完全了解lncRNA,mRNA和circRNA在OA中的表达特征和功能。
目的:本研究旨在鉴定OA中差异表达的基因。
方法:本研究进行了RNA测序,以鉴定OA中差异表达的基因。本体(GO)分析用于分析分子功能和生物过程。KEGG途径分析用于在生物学途径中进行差异表达的lncRNA。
结果:分层聚类显示总共943个mRNA,518个lncRNA和300个circRNA,与正常样品相比,它们在OA中失调。此外,我们构建了差异表达的mRNA介导的蛋白质-蛋白质相互作用网络,差异表达的lncRNA介导的反式调节网络和竞争性内源RNA(ceRNA),以揭示OA中这些基因之间的相互作用。生物信息学分析显示,这些失调的基因参与调节多个生物学过程,例如伤口愈合,骨化的负调节,姐妹染色单体凝聚,白介素-1α产生的正调节,钠离子跨膜转运,细胞迁移的正调节以及炎症反应的负调控。据我们所知,
结论:这项研究提供了新颖的信息,以验证这些差异表达的RNAs可能是OA中的生物标志物和靶标。



























 京公网安备 11010802027423号
京公网安备 11010802027423号