ISPRS Journal of Photogrammetry and Remote Sensing ( IF 10.6 ) Pub Date : 2021-02-06 , DOI: 10.1016/j.isprsjprs.2021.01.017 Martin Danner , Katja Berger , Matthias Wocher , Wolfram Mauser , Tobias Hank
|
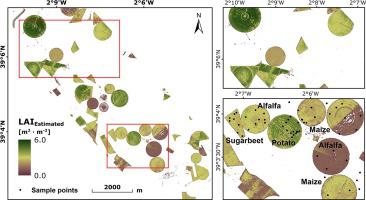
With an upcoming unprecedented stream of imaging spectroscopy data, there is a rising need for tools and software applications exploiting the spectral possibilities to extract relevant information on an operational basis. In this study, we investigate the potential of a scientific processor designed to quantify biophysical and biochemical crop traits from spectroscopic imagery of the upcoming Environmental Mapping and Analysis Program (EnMAP) satellite. Said processor relies on a hybrid retrieval workflow executing pre-trained machine learning regression models fast and efficiently based on training data from a lookup table of synthetic vegetation spectra and their associated parameterization of the well-known radiative transfer model (RTM) PROSAIL. The established models provide spatial information about leaf area index (LAI), average leaf inclination angle (ALIA), leaf chlorophyll content (Cab) and leaf mass per area (Cm). In contrast to using site-specific training data, the approach facilitates a universal application without the need to integrate a priori information into the processor. Four machine learning algorithms, namely artificial neural networks (ANN), random forest regression (RFR), support vector machine regression (SVR), and Gaussian process regression (GPR), were found to estimate biophysical and biochemical variables of unseen targets with high performance (relative error scores < 10%). ANNs excelled in terms of accuracy, model size and execution time when the 242 spectral bands were transformed into 15 principal components, the signals of which were scaled by a z-transformation. Validation using in situ data from the SPARC03 Barrax campaign dataset revealed an overall good estimation of measured functional traits, for instance for LAI with root mean squared error (RMSE) of 0.81 m2 m−2, and for Cab RMSE of 6.2 µg cm−2 with the ANN model. Moreover, both crop traits could be successfully mapped using a pseudo-EnMAP scene revealing plausible within-field patterns. Conformity with LAI output of the SNAP biophysical processor was found especially for grassland and maize in the vegetative stages. Based on these findings, ANN models are considered the best choice for implementation of a hybrid retrieval workflow within the context of operational agricultural crop traits monitoring from future satellite imaging spectroscopy.
中文翻译:

基于RTM的高效机器学习回归算法训练,以量化农作物的生物物理和生化特性
随着即将到来的前所未有的成像光谱数据流,对工具和软件应用的需求不断增长,这些工具和软件应利用光谱的可能性在操作基础上提取相关信息。在这项研究中,我们将研究一种科学处理器的潜力,该处理器旨在从即将到来的环境制图和分析计划(EnMAP)卫星的光谱图像中量化生物物理和生化作物的性状。所述处理器依赖于混合检索工作流,该混合检索工作流基于来自合成植物光谱查找表的训练数据及其众所周知的辐射传递模型(RTM)PROSAIL的相关参数化,快速有效地执行预训练的机器学习回归模型。建立的模型提供有关叶面积指数(LAI)的空间信息,ab)和单位面积的叶片质量(C m)。与使用特定于站点的培训数据相反,该方法无需将先验信息集成到处理器中即可促进通用应用程序。发现了四种机器学习算法,即人工神经网络(ANN),随机森林回归(RFR),支持向量机回归(SVR)和高斯过程回归(GPR),可高效估计未见目标的生物物理和生化变量(相对错误分数<10%)。当将242个光谱带转换为15个主要分量时,人工神经网络在准确性,模型大小和执行时间方面均表现出色,其信号通过z变换进行缩放。使用来自SPARC03 Barrax广告活动数据集的原位数据进行的验证显示,对所测功能性状的总体评价不错,2 m -2,对于Aab模型,C ab RMSE为6.2 µg cm -2。此外,可以使用伪EnMAP场景成功绘制两个作物性状,从而揭示合理的田间模式。发现与SNAP生物物理处理器的LAI输出一致,特别是在营养阶段的草地和玉米。基于这些发现,ANN模型被认为是在未来的卫星成像光谱法对农业农作物性状进行监测的前提下,实施混合检索工作流程的最佳选择。











































 京公网安备 11010802027423号
京公网安备 11010802027423号