Fungal Biology ( IF 2.9 ) Pub Date : 2021-02-04 , DOI: 10.1016/j.funbio.2021.01.006 Sandra Hilário 1 , Liliana Santos 1 , Artur Alves 1
|
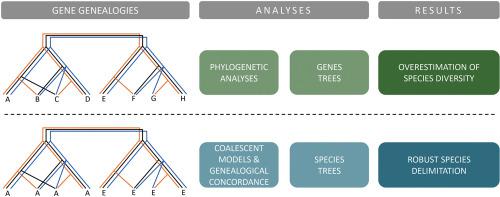
Delimitation of species boundaries within the fungal genus Diaporthe has been challenging, but the analyses of combined multilocus DNA sequences has become an important tool to infer phylogenetic relationships and to circumscribe species. However, analyses of congruence between individual gene genealogies and the application of the genealogical concordance principle have been somehow overlooked. We noted that a group of species including D. amygdali, D. garethjonesii, D. sterilis, D. kadsurae, D. ternstroemia, D. ovoicicola, D. fusicola, D. chongqingensis and D. mediterranea, commonly known as D. amygdali complex, occupy a monophyletic clade in Diaporthe phylogenies but the limits of all species within the complex are not entirely clear. To assess the boundaries of species within this complex we employed the Genealogical Concordance Phylogenetic Species Recognition principle (GCPSR) and coalescence-based models: General Mixed Yule-Coalescent (GMYC) and Poisson Tree Processes (PTP). The incongruence detected between individual gene phylogenies, as well as the results of coalescent methods do not support the recognition of lineages within the complex as distinct species. Moreover, results support the absence of reproductive isolation and barriers to gene flow in this complex, thus providing further evidence that the D. amygdali species complex constitutes a single species. This study highlights the relevance of the application of the GCPSR principle, showing that concatenation analysis of multilocus DNA sequences, although being a powerful tool, might lead to an erroneous definition of species limits. Additionally, it further shows that coalescent methods are useful tools to assist in a more robust delimitation of species boundaries in the genus Diaporthe.
中文翻译:

Diaporthe amygdali,一个物种复合体还是一个复杂的物种?
在真菌属Diaporthe内划定物种边界一直具有挑战性,但组合多位点 DNA 序列的分析已成为推断系统发育关系和限制物种的重要工具。然而,个体基因谱系之间一致性的分析和谱系一致性原则的应用在某种程度上被忽视了。我们注意到一组物种包括D. amygdali , D. garethjonesii , D. sterilis , D. kadsurae , D. ternstroemia , D. ovoicicola , D. fusicola , D. chongqingensis和D. mediterranea,通常称为D. amygdali复合体,在Diaporthe系统发育中占据单系进化枝,但复合体中所有物种的界限并不完全清楚。为了评估该复合体中物种的边界,我们采用了系谱一致性系统发育物种识别原理 (GCPSR) 和基于合并的模型:通用混合圣诞合并 (GMYC) 和泊松树过程 (PTP)。在单个基因系统发育之间检测到的不一致以及合并方法的结果不支持将复杂内的谱系识别为不同的物种。此外,结果支持该复合体中不存在生殖隔离和基因流动障碍,从而提供进一步的证据表明D. amygdali物种复合体构成单一物种。这项研究强调了 GCPSR 原理应用的相关性,表明多位点 DNA 序列的串联分析虽然是一个强大的工具,但可能会导致物种限制的错误定义。此外,它还进一步表明,聚结方法是有助于更有效地划定Diaporthe属物种边界的有用工具。










































 京公网安备 11010802027423号
京公网安备 11010802027423号