当前位置:
X-MOL 学术
›
Hum. Brain Mapp.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
High resolution automated labeling of the hippocampus and amygdala using a 3D convolutional neural network trained on whole brain 700 μm isotropic 7T MP2RAGE MRI
Human Brain Mapping ( IF 3.5 ) Pub Date : 2021-01-25 , DOI: 10.1002/hbm.25348 Heath R Pardoe 1 , Arun Raj Antony 2 , Hoby Hetherington 2 , Anto I Bagić 2 , Timothy M Shepherd 3 , Daniel Friedman 1 , Orrin Devinsky 1 , Jullie Pan 2
Human Brain Mapping ( IF 3.5 ) Pub Date : 2021-01-25 , DOI: 10.1002/hbm.25348 Heath R Pardoe 1 , Arun Raj Antony 2 , Hoby Hetherington 2 , Anto I Bagić 2 , Timothy M Shepherd 3 , Daniel Friedman 1 , Orrin Devinsky 1 , Jullie Pan 2
Affiliation
|
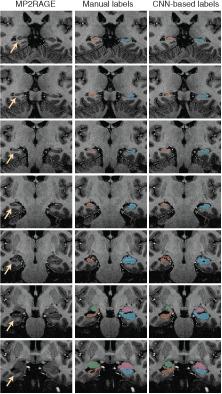
Image labeling using convolutional neural networks (CNNs) are a template‐free alternative to traditional morphometric techniques. We trained a 3D deep CNN to label the hippocampus and amygdala on whole brain 700 μm isotropic 3D MP2RAGE MRI acquired at 7T. Manual labels of the hippocampus and amygdala were used to (i) train the predictive model and (ii) evaluate performance of the model when applied to new scans. Healthy controls and individuals with epilepsy were included in our analyses. Twenty‐one healthy controls and sixteen individuals with epilepsy were included in the study. We utilized the recently developed DeepMedic software to train a CNN to label the hippocampus and amygdala based on manual labels. Performance was evaluated by measuring the dice similarity coefficient (DSC) between CNN‐based and manual labels. A leave‐one‐out cross validation scheme was used. CNN‐based and manual volume estimates were compared for the left and right hippocampus and amygdala in healthy controls and epilepsy cases. The CNN‐based technique successfully labeled the hippocampus and amygdala in all cases. Mean DSC = 0.88 ± 0.03 for the hippocampus and 0.8 ± 0.06 for the amygdala. CNN‐based labeling was independent of epilepsy diagnosis in our sample (p = .91). CNN‐based volume estimates were highly correlated with manual volume estimates in epilepsy cases and controls. CNNs can label the hippocampus and amygdala on native sub‐mm resolution MP2RAGE 7T MRI. Our findings suggest deep learning techniques can advance development of morphometric analysis techniques for high field strength, high spatial resolution brain MRI.
中文翻译:

使用在全脑 700 μm 各向同性 7T MP2RAGE MRI 上训练的 3D 卷积神经网络对海马体和杏仁核进行高分辨率自动标记
使用卷积神经网络 (CNN) 的图像标记是传统形态测量技术的无模板替代方案。我们训练了一个 3D 深度 CNN,以在 7T 采集的全脑 700 μm 各向同性 3D MP2RAGE MRI 上标记海马体和杏仁核。海马体和杏仁核的手动标签用于 (i) 训练预测模型和 (ii) 在应用于新扫描时评估模型的性能。我们的分析中包括健康对照和癫痫患者。研究包括 21 名健康对照者和 16 名癫痫患者。我们利用最近开发的 DeepMedic 软件来训练 CNN,以根据手动标签来标记海马体和杏仁核。通过测量基于 CNN 的标签和手动标签之间的骰子相似系数 (DSC) 来评估性能。使用了留一法交叉验证方案。在健康对照组和癫痫病例中,比较了基于 CNN 和手动体积估计的左右海马和杏仁核。基于 CNN 的技术在所有情况下都成功地标记了海马体和杏仁核。海马的平均 DSC = 0.88 ± 0.03,杏仁核的平均 DSC = 0.8 ± 0.06。在我们的样本中,基于 CNN 的标记与癫痫诊断无关(p = .91)。在癫痫病例和对照组中,基于 CNN 的体积估计与手动体积估计高度相关。CNN 可以在原生亚毫米分辨率 MP2RAGE 7T MRI 上标记海马体和杏仁核。我们的研究结果表明,深度学习技术可以促进高场强、高空间分辨率脑 MRI 的形态分析技术的发展。
更新日期:2021-01-25
中文翻译:

使用在全脑 700 μm 各向同性 7T MP2RAGE MRI 上训练的 3D 卷积神经网络对海马体和杏仁核进行高分辨率自动标记
使用卷积神经网络 (CNN) 的图像标记是传统形态测量技术的无模板替代方案。我们训练了一个 3D 深度 CNN,以在 7T 采集的全脑 700 μm 各向同性 3D MP2RAGE MRI 上标记海马体和杏仁核。海马体和杏仁核的手动标签用于 (i) 训练预测模型和 (ii) 在应用于新扫描时评估模型的性能。我们的分析中包括健康对照和癫痫患者。研究包括 21 名健康对照者和 16 名癫痫患者。我们利用最近开发的 DeepMedic 软件来训练 CNN,以根据手动标签来标记海马体和杏仁核。通过测量基于 CNN 的标签和手动标签之间的骰子相似系数 (DSC) 来评估性能。使用了留一法交叉验证方案。在健康对照组和癫痫病例中,比较了基于 CNN 和手动体积估计的左右海马和杏仁核。基于 CNN 的技术在所有情况下都成功地标记了海马体和杏仁核。海马的平均 DSC = 0.88 ± 0.03,杏仁核的平均 DSC = 0.8 ± 0.06。在我们的样本中,基于 CNN 的标记与癫痫诊断无关(p = .91)。在癫痫病例和对照组中,基于 CNN 的体积估计与手动体积估计高度相关。CNN 可以在原生亚毫米分辨率 MP2RAGE 7T MRI 上标记海马体和杏仁核。我们的研究结果表明,深度学习技术可以促进高场强、高空间分辨率脑 MRI 的形态分析技术的发展。











































 京公网安备 11010802027423号
京公网安备 11010802027423号