当前位置:
X-MOL 学术
›
Environ. Sci.: Water Res. Technol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the U.S.
Environmental Science: Water Research & Technology ( IF 3.5 ) Pub Date : 2021-1-20 , DOI: 10.1039/d0ew00946f Brian M Pecson 1 , Emily Darby 1 , Charles N Haas 2 , Yamrot M Amha 3 , Mitchel Bartolo 4 , Richard Danielson 5 , Yeggie Dearborn 5 , George Di Giovanni 6 , Christobel Ferguson 7 , Stephanie Fevig 7 , Erica Gaddis 8 , Donald Gray 9 , George Lukasik 10 , Bonnie Mull 10 , Liana Olivas 3 , Adam Olivieri 11 , Yan Qu 3 ,
Environmental Science: Water Research & Technology ( IF 3.5 ) Pub Date : 2021-1-20 , DOI: 10.1039/d0ew00946f Brian M Pecson 1 , Emily Darby 1 , Charles N Haas 2 , Yamrot M Amha 3 , Mitchel Bartolo 4 , Richard Danielson 5 , Yeggie Dearborn 5 , George Di Giovanni 6 , Christobel Ferguson 7 , Stephanie Fevig 7 , Erica Gaddis 8 , Donald Gray 9 , George Lukasik 10 , Bonnie Mull 10 , Liana Olivas 3 , Adam Olivieri 11 , Yan Qu 3 ,
Affiliation
|
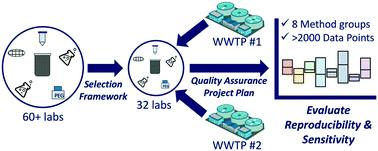
In response to COVID-19, the international water community rapidly developed methods to quantify the SARS-CoV-2 genetic signal in untreated wastewater. Wastewater surveillance using such methods has the potential to complement clinical testing in assessing community health. This interlaboratory assessment evaluated the reproducibility and sensitivity of 36 standard operating procedures (SOPs), divided into eight method groups based on sample concentration approach and whether solids were removed. Two raw wastewater samples were collected in August 2020, amended with a matrix spike (betacoronavirus OC43), and distributed to 32 laboratories across the U.S. Replicate samples analyzed in accordance with the project's quality assurance plan showed high reproducibility across the 36 SOPs: 80% of the recovery-corrected results fell within a band of ±1.15 log10 genome copies per L with higher reproducibility observed within a single SOP (standard deviation of 0.13 log10). The inclusion of a solids removal step and the selection of a concentration method did not show a clear, systematic impact on the recovery-corrected results. Other methodological variations (e.g., pasteurization, primer set selection, and use of RT-qPCR or RT-dPCR platforms) generally resulted in small differences compared to other sources of variability. These findings suggest that a variety of methods are capable of producing reproducible results, though the same SOP or laboratory should be selected to track SARS-CoV-2 trends at a given facility. The methods showed a 7 log10 range of recovery efficiency and limit of detection highlighting the importance of recovery correction and the need to consider method sensitivity when selecting methods for wastewater surveillance.
中文翻译:

36 种量化原废水中 SARS-CoV-2 基因信号的方法的重现性和灵敏度:美国实验室间方法评估的结果
为了应对 COVID-19,国际水界迅速开发了量化未经处理废水中 SARS-CoV-2 基因信号的方法。使用此类方法进行废水监测有可能补充评估社区健康的临床测试。该实验室间评估评估了 36 个标准操作程序 (SOP) 的再现性和灵敏度,根据样品浓度方法和固体是否被去除分为八个方法组。 2020 年 8 月收集了两份原废水样本,并用基质尖峰(β冠状病毒 OC43)进行了修正,并分发给美国各地的 32 个实验室。根据项目的质量保证计划分析的重复样本在 36 个 SOP 中显示出高重现性:80%恢复校正结果落在每升 ±1.15 log 10基因组拷贝的范围内,在单个 SOP 内观察到更高的重现性(标准差为 0.13 log 10 )。固体去除步骤和浓缩方法的选择并未显示出对回收率校正结果的明显、系统性影响。与其他变异来源相比,其他方法学变化(例如巴氏灭菌、引物组选择以及 RT-qPCR 或 RT-dPCR 平台的使用)通常会导致较小的差异。这些发现表明,尽管应选择相同的 SOP 或实验室来跟踪给定设施中的 SARS-CoV-2 趋势,但多种方法都能够产生可重复的结果。 该方法显示了 7 log 10的回收率范围和检测限,突出了回收率校正的重要性以及在选择废水监测方法时需要考虑方法灵敏度。
更新日期:2021-01-22
中文翻译:

36 种量化原废水中 SARS-CoV-2 基因信号的方法的重现性和灵敏度:美国实验室间方法评估的结果
为了应对 COVID-19,国际水界迅速开发了量化未经处理废水中 SARS-CoV-2 基因信号的方法。使用此类方法进行废水监测有可能补充评估社区健康的临床测试。该实验室间评估评估了 36 个标准操作程序 (SOP) 的再现性和灵敏度,根据样品浓度方法和固体是否被去除分为八个方法组。 2020 年 8 月收集了两份原废水样本,并用基质尖峰(β冠状病毒 OC43)进行了修正,并分发给美国各地的 32 个实验室。根据项目的质量保证计划分析的重复样本在 36 个 SOP 中显示出高重现性:80%恢复校正结果落在每升 ±1.15 log 10基因组拷贝的范围内,在单个 SOP 内观察到更高的重现性(标准差为 0.13 log 10 )。固体去除步骤和浓缩方法的选择并未显示出对回收率校正结果的明显、系统性影响。与其他变异来源相比,其他方法学变化(例如巴氏灭菌、引物组选择以及 RT-qPCR 或 RT-dPCR 平台的使用)通常会导致较小的差异。这些发现表明,尽管应选择相同的 SOP 或实验室来跟踪给定设施中的 SARS-CoV-2 趋势,但多种方法都能够产生可重复的结果。 该方法显示了 7 log 10的回收率范围和检测限,突出了回收率校正的重要性以及在选择废水监测方法时需要考虑方法灵敏度。









































 京公网安备 11010802027423号
京公网安备 11010802027423号