当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Systematic Identification of Protein Phosphorylation-Mediated Interactions
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2021-01-21 , DOI: 10.1021/acs.jproteome.0c00750 Brendan M Floyd 1 , Kevin Drew 1 , Edward M Marcotte 1
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2021-01-21 , DOI: 10.1021/acs.jproteome.0c00750 Brendan M Floyd 1 , Kevin Drew 1 , Edward M Marcotte 1
Affiliation
|
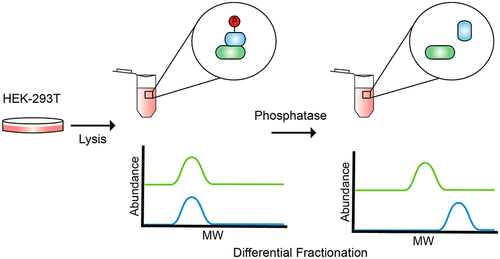
Protein phosphorylation is a key regulatory mechanism involved in nearly every eukaryotic cellular process. Increasingly sensitive mass spectrometry approaches have identified hundreds of thousands of phosphorylation sites, but the functions of a vast majority of these sites remain unknown, with fewer than 5% of sites currently assigned a function. To increase our understanding of functional protein phosphorylation we developed an approach (phospho-DIFFRAC) for identifying the phosphorylation-dependence of protein assemblies in a systematic manner. A combination of nonspecific protein phosphatase treatment, size-exclusion chromatography, and mass spectrometry allowed us to identify changes in protein interactions after the removal of phosphate modifications. With this approach we were able to identify 316 proteins involved in phosphorylation-sensitive interactions. We recovered known phosphorylation-dependent interactors such as the FACT complex and spliceosome, as well as identified novel interactions such as the tripeptidyl peptidase TPP2 and the supraspliceosome component ZRANB2. More generally, we find phosphorylation-dependent interactors to be strongly enriched for RNA-binding proteins, providing new insight into the role of phosphorylation in RNA binding. By searching directly for phosphorylated amino acid residues in mass spectrometry data, we identified the likely regulatory phosphosites on ZRANB2 and FACT complex subunit SSRP1. This study provides both a method and resource for obtaining a better understanding of the role of phosphorylation in native macromolecular assemblies. All mass spectrometry data are available through PRIDE (accession #PXD021422).
中文翻译:

蛋白质磷酸化介导的相互作用的系统鉴定
蛋白质磷酸化是涉及几乎所有真核细胞过程的关键调节机制。越来越敏感的质谱方法已经确定了数十万个磷酸化位点,但其中绝大多数位点的功能仍然未知,目前只有不到 5% 的位点具有功能。为了增加我们对功能性蛋白质磷酸化的理解,我们开发了一种方法 (phospho-DIFFRAC),用于系统地识别蛋白质组装体的磷酸化依赖性。非特异性蛋白磷酸酶处理、尺寸排阻色谱法和质谱法的组合使我们能够识别去除磷酸盐修饰后蛋白质相互作用的变化。通过这种方法,我们能够识别出 316 种参与磷酸化敏感相互作用的蛋白质。我们恢复了已知的磷酸化依赖性相互作用物,例如 FACT 复合物和剪接体,以及确定的新型相互作用,例如三肽基肽酶 TPP2 和超剪接体成分 ZRANB2。更一般地说,我们发现磷酸化依赖性相互作用物强烈富集 RNA 结合蛋白,为磷酸化在 RNA 结合中的作用提供了新的见解。通过直接搜索质谱数据中的磷酸化氨基酸残基,我们确定了 ZRANB2 和 FACT 复合物亚基 SSRP1 上可能的调节磷酸位点。本研究为更好地了解磷酸化在天然大分子组装中的作用提供了一种方法和资源。
更新日期:2021-02-05
中文翻译:

蛋白质磷酸化介导的相互作用的系统鉴定
蛋白质磷酸化是涉及几乎所有真核细胞过程的关键调节机制。越来越敏感的质谱方法已经确定了数十万个磷酸化位点,但其中绝大多数位点的功能仍然未知,目前只有不到 5% 的位点具有功能。为了增加我们对功能性蛋白质磷酸化的理解,我们开发了一种方法 (phospho-DIFFRAC),用于系统地识别蛋白质组装体的磷酸化依赖性。非特异性蛋白磷酸酶处理、尺寸排阻色谱法和质谱法的组合使我们能够识别去除磷酸盐修饰后蛋白质相互作用的变化。通过这种方法,我们能够识别出 316 种参与磷酸化敏感相互作用的蛋白质。我们恢复了已知的磷酸化依赖性相互作用物,例如 FACT 复合物和剪接体,以及确定的新型相互作用,例如三肽基肽酶 TPP2 和超剪接体成分 ZRANB2。更一般地说,我们发现磷酸化依赖性相互作用物强烈富集 RNA 结合蛋白,为磷酸化在 RNA 结合中的作用提供了新的见解。通过直接搜索质谱数据中的磷酸化氨基酸残基,我们确定了 ZRANB2 和 FACT 复合物亚基 SSRP1 上可能的调节磷酸位点。本研究为更好地了解磷酸化在天然大分子组装中的作用提供了一种方法和资源。









































 京公网安备 11010802027423号
京公网安备 11010802027423号