Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Using Bibliometric Analysis and Machine Learning to Identify Compounds Binding to Sialidase-1
ACS Omega ( IF 3.7 ) Pub Date : 2021-01-20 , DOI: 10.1021/acsomega.0c05591 Jennifer J Klein 1 , Nancy C Baker 2 , Daniel H Foil 1 , Kimberley M Zorn 1 , Fabio Urbina 1 , Ana C Puhl 1 , Sean Ekins 1
ACS Omega ( IF 3.7 ) Pub Date : 2021-01-20 , DOI: 10.1021/acsomega.0c05591 Jennifer J Klein 1 , Nancy C Baker 2 , Daniel H Foil 1 , Kimberley M Zorn 1 , Fabio Urbina 1 , Ana C Puhl 1 , Sean Ekins 1
Affiliation
|
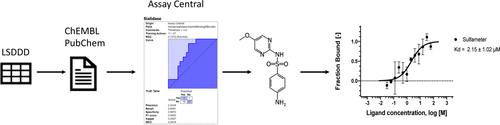
Rare diseases impact hundreds of millions of individuals worldwide. However, few therapies exist to treat the rare disease population because financial resources are limited, the number of patients affected is low, bioactivity data is often nonexistent, and very few animal models exist to support preclinical development efforts. Sialidosis is an ultrarare lysosomal storage disorder in which mutations in the NEU1 gene result in the deficiency of the lysosomal enzyme sialidase-1. This enzyme catalyzes the removal of sialic acid moieties from glycoproteins and glycolipids. Therefore, the defective or deficient protein leads to the buildup of sialylated glycoproteins as well as several characteristic symptoms of sialidosis including visual impairment, ataxia, hepatomegaly, dysostosis multiplex, and developmental delay. In this study, we used a bibliometric tool to generate links between lysosomal storage disease (LSD) targets and existing bioactivity data that could be curated in order to build machine learning models and screen compounds in silico. We focused on sialidase as an example, and we used the data curated from the literature to build a Bayesian model which was then used to score compound libraries and rank these molecules for in vitro testing. Two compounds were identified from in vitro testing using microscale thermophoresis, namely sulfameter (Kd 2.15 ± 1.02 μM) and mexenone (Kd 8.88 ± 4.02 μM), which validated our approach to identifying new molecules binding to this protein, which could represent possible drug candidates that can be evaluated further as potential chaperones for this ultrarare lysosomal disease for which there is currently no treatment. Combining bibliometric and machine learning approaches has the ability to assist in curating small molecule data and model building, respectively, for rare disease drug discovery. This approach also has the capability to identify new compounds that are potential drug candidates.
中文翻译:

使用文献计量分析和机器学习来识别与 Sialidase-1 结合的化合物
罕见疾病影响着全世界数亿人。然而,治疗罕见疾病人群的疗法很少,因为财政资源有限,受影响的患者数量很少,生物活性数据通常不存在,并且支持临床前开发工作的动物模型很少。唾液酸贮积症是一种极为罕见的溶酶体贮积症,NEU1 基因突变导致溶酶体唾液酸酶-1 缺乏。该酶催化从糖蛋白和糖脂中去除唾液酸部分。因此,有缺陷或缺乏的蛋白质会导致唾液酸化糖蛋白的积聚以及唾液酸贮积症的几种特征性症状,包括视力障碍、共济失调、肝肿大、多发性骨发育不全和发育迟缓。在这项研究中,我们使用文献计量工具来生成溶酶体贮积病 (LSD) 目标与现有生物活性数据之间的联系,这些数据可以进行整理,以便构建机器学习模型并在计算机中筛选化合物。我们以唾液酸酶为例,使用文献中整理的数据构建贝叶斯模型,然后使用该模型对化合物库进行评分并对这些分子进行排名以进行体外测试。使用微量热泳法在体外测试中鉴定出两种化合物,即磺胺计( K d 2.15 ± 1.02 μM)和美塞酮( K d 8.88 ± 4.02 μM),这验证了我们识别与该蛋白质结合的新分子的方法,这可能代表了可能的结果。可以进一步评估候选药物作为这种目前尚无治疗方法的极其罕见的溶酶体疾病的潜在伴侣。 结合文献计量和机器学习方法能够分别帮助管理小分子数据和模型构建,以用于罕见疾病药物的发现。这种方法还能够识别作为潜在候选药物的新化合物。
更新日期:2021-02-02
中文翻译:

使用文献计量分析和机器学习来识别与 Sialidase-1 结合的化合物
罕见疾病影响着全世界数亿人。然而,治疗罕见疾病人群的疗法很少,因为财政资源有限,受影响的患者数量很少,生物活性数据通常不存在,并且支持临床前开发工作的动物模型很少。唾液酸贮积症是一种极为罕见的溶酶体贮积症,NEU1 基因突变导致溶酶体唾液酸酶-1 缺乏。该酶催化从糖蛋白和糖脂中去除唾液酸部分。因此,有缺陷或缺乏的蛋白质会导致唾液酸化糖蛋白的积聚以及唾液酸贮积症的几种特征性症状,包括视力障碍、共济失调、肝肿大、多发性骨发育不全和发育迟缓。在这项研究中,我们使用文献计量工具来生成溶酶体贮积病 (LSD) 目标与现有生物活性数据之间的联系,这些数据可以进行整理,以便构建机器学习模型并在计算机中筛选化合物。我们以唾液酸酶为例,使用文献中整理的数据构建贝叶斯模型,然后使用该模型对化合物库进行评分并对这些分子进行排名以进行体外测试。使用微量热泳法在体外测试中鉴定出两种化合物,即磺胺计( K d 2.15 ± 1.02 μM)和美塞酮( K d 8.88 ± 4.02 μM),这验证了我们识别与该蛋白质结合的新分子的方法,这可能代表了可能的结果。可以进一步评估候选药物作为这种目前尚无治疗方法的极其罕见的溶酶体疾病的潜在伴侣。 结合文献计量和机器学习方法能够分别帮助管理小分子数据和模型构建,以用于罕见疾病药物的发现。这种方法还能够识别作为潜在候选药物的新化合物。











































 京公网安备 11010802027423号
京公网安备 11010802027423号