当前位置:
X-MOL 学术
›
Biopolymers
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Identifying DNA ‐binding proteins based on multi‐features and LASSO feature selection
Biopolymers ( IF 3.2 ) Pub Date : 2021-01-21 , DOI: 10.1002/bip.23419 Shengli Zhang 1 , Fu Zhu 1 , Qianhao Yu 2 , Xiaoyue Zhu 3
Biopolymers ( IF 3.2 ) Pub Date : 2021-01-21 , DOI: 10.1002/bip.23419 Shengli Zhang 1 , Fu Zhu 1 , Qianhao Yu 2 , Xiaoyue Zhu 3
Affiliation
|
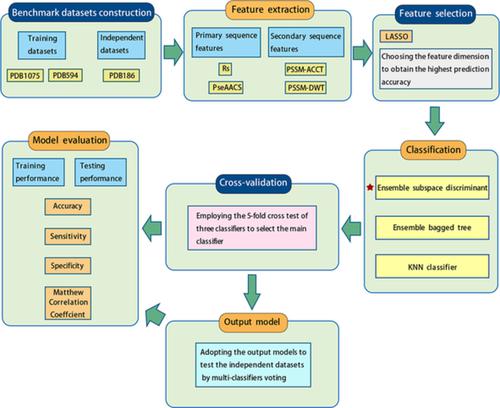
DNA-binding proteins perform an indispensable function in the maintenance and processing of genetic information and are inefficiently identified by traditional experimental methods due to their huge quantities. On the contrary, machine learning methods as an emerging technique demonstrate satisfactory speed and accuracy when used to study these molecules. This work focuses on extracting four different features from primary and secondary sequence features: Reduced sequence and index-vectors (RS), Pseudo-amino acid components (PseAACS), Position-specific scoring matrix-Auto Cross Covariance Transform (PSSM-ACCT), and Position-specific scoring matrix-Discrete Wavelet Transform (PSSM-DWT). Using the LASSO dimension reduction method, we experiment on the combination of feature submodels to obtain the optimized number of top rank features. These features are respectively input into the training Ensemble subspace discriminant, Ensemble bagged tree and KNN to predict the DNA-binding proteins. Three different datasets, PDB594, PDB1075, and PDB186, are adopted to evaluate the performance of the as-proposed approach in this work. The PDB1075 and PDB594 datasets are adopted for the five-fold cross-validation, and the PDB186 is used for the independent experiment. In the five-fold cross-validation, both the PDB1075 and PDB594 show extremely high accuracy, reaching 86.98% and 88.9% by Ensemble subspace discriminant, respectively. The accuracy of independent experiment by multi-classifiers voting is 83.33%, which suggests that the methodology proposed in this work is capable of predicting DNA-binding proteins effectively.
中文翻译:

基于多特征和LASSO特征选择识别DNA结合蛋白
DNA结合蛋白在遗传信息的维护和处理中发挥着不可或缺的作用,但由于数量庞大,传统实验方法无法有效识别。相反,机器学习方法作为一种新兴技术在用于研究这些分子时表现出令人满意的速度和准确性。这项工作的重点是从一级和二级序列特征中提取四种不同的特征:缩减序列和索引向量 (RS)、伪氨基酸成分 (PseAACS)、特定位置评分矩阵-自动交叉协方差变换 (PSSM-ACCT)、和位置特定的评分矩阵 - 离散小波变换 (PSSM-DWT)。使用 LASSO 降维方法,我们对特征子模型的组合进行了实验,以获得优化的顶级特征数量。这些特征分别输入到训练 Ensemble 子空间判别器、Ensemble 袋装树和 KNN 中以预测 DNA 结合蛋白。采用三个不同的数据集 PDB594、PDB1075 和 PDB186 来评估本文提出的方法的性能。五重交叉验证采用PDB1075和PDB594数据集,独立实验采用PDB186数据集。在五重交叉验证中,PDB1075 和 PDB594 都表现出极高的准确率,Ensemble 子空间判别分别达到 86.98% 和 88.9%。多分类器投票的独立实验准确率为 83.33%,表明本文提出的方法能够有效预测 DNA 结合蛋白。
更新日期:2021-01-21
中文翻译:

基于多特征和LASSO特征选择识别DNA结合蛋白
DNA结合蛋白在遗传信息的维护和处理中发挥着不可或缺的作用,但由于数量庞大,传统实验方法无法有效识别。相反,机器学习方法作为一种新兴技术在用于研究这些分子时表现出令人满意的速度和准确性。这项工作的重点是从一级和二级序列特征中提取四种不同的特征:缩减序列和索引向量 (RS)、伪氨基酸成分 (PseAACS)、特定位置评分矩阵-自动交叉协方差变换 (PSSM-ACCT)、和位置特定的评分矩阵 - 离散小波变换 (PSSM-DWT)。使用 LASSO 降维方法,我们对特征子模型的组合进行了实验,以获得优化的顶级特征数量。这些特征分别输入到训练 Ensemble 子空间判别器、Ensemble 袋装树和 KNN 中以预测 DNA 结合蛋白。采用三个不同的数据集 PDB594、PDB1075 和 PDB186 来评估本文提出的方法的性能。五重交叉验证采用PDB1075和PDB594数据集,独立实验采用PDB186数据集。在五重交叉验证中,PDB1075 和 PDB594 都表现出极高的准确率,Ensemble 子空间判别分别达到 86.98% 和 88.9%。多分类器投票的独立实验准确率为 83.33%,表明本文提出的方法能够有效预测 DNA 结合蛋白。









































 京公网安备 11010802027423号
京公网安备 11010802027423号