当前位置:
X-MOL 学术
›
Chem. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Snapshotting the transient conformations and tracing the multiple pathways of single peptide folding using a solid-state nanopore
Chemical Science ( IF 7.6 ) Pub Date : 2021-1-4 , DOI: 10.1039/d0sc06106a Shao-Chuang Liu 1, 2 , Yi-Lun Ying 1, 2 , Wei-Hua Li 3 , Yong-Jing Wan 4 , Yi-Tao Long 1, 2
Chemical Science ( IF 7.6 ) Pub Date : 2021-1-4 , DOI: 10.1039/d0sc06106a Shao-Chuang Liu 1, 2 , Yi-Lun Ying 1, 2 , Wei-Hua Li 3 , Yong-Jing Wan 4 , Yi-Tao Long 1, 2
Affiliation
|
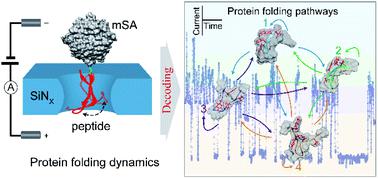
A fundamental question relating to protein folding/unfolding is the time evolution of the folding of a protein into its precisely defined native structure. The proper identification of transition conformations is essential for accurately describing the dynamic protein folding/unfolding pathways. Owing to the rapid transitions and sub-nm conformation differences involved, the acquisition of the transient conformations and dynamics of proteins is difficult due to limited instrumental resolution. Using the electrochemical confinement effect of a solid-state nanopore, we were able to snapshot the transient conformations and trace the multiple transition pathways of a single peptide inside a nanopore. By combining the results with a Markov chain model, this new single-molecule technique is applied to clarify the transition pathways of the β-hairpin peptide, which shows nonequilibrium fluctuations among several blockage current stages. This method enables the high-throughput investigation of transition pathways experimentally to access previously obscure peptide dynamics, which is significant for understanding the folding/unfolding mechanisms and misfolding of peptides or proteins.
中文翻译:

使用固态纳米孔捕捉瞬时构象并追踪单肽折叠的多种途径
与蛋白质折叠/展开有关的一个基本问题是蛋白质折叠成其精确定义的天然结构的时间演变。正确识别过渡构象对于准确描述动态蛋白质折叠/展开路径至关重要。由于涉及到快速转变和亚纳米构象差异,由于仪器分辨率有限,难以获得蛋白质的瞬时构象和动力学。利用固态纳米孔的电化学限制效应,我们能够捕捉瞬态构象并追踪纳米孔内单个肽的多个过渡途径。通过将结果与马尔可夫链模型相结合,这项新的单分子技术被用于阐明β-发夹肽的过渡途径,该途径显示了多个阻断电流阶段之间的非平衡波动。该方法可以通过实验对过渡途径进行高通量研究,以获取先前难以理解的肽动力学,这对于理解肽或蛋白质的折叠/解折叠机制和错误折叠具有重要意义。
更新日期:2021-01-20
中文翻译:

使用固态纳米孔捕捉瞬时构象并追踪单肽折叠的多种途径
与蛋白质折叠/展开有关的一个基本问题是蛋白质折叠成其精确定义的天然结构的时间演变。正确识别过渡构象对于准确描述动态蛋白质折叠/展开路径至关重要。由于涉及到快速转变和亚纳米构象差异,由于仪器分辨率有限,难以获得蛋白质的瞬时构象和动力学。利用固态纳米孔的电化学限制效应,我们能够捕捉瞬态构象并追踪纳米孔内单个肽的多个过渡途径。通过将结果与马尔可夫链模型相结合,这项新的单分子技术被用于阐明β-发夹肽的过渡途径,该途径显示了多个阻断电流阶段之间的非平衡波动。该方法可以通过实验对过渡途径进行高通量研究,以获取先前难以理解的肽动力学,这对于理解肽或蛋白质的折叠/解折叠机制和错误折叠具有重要意义。









































 京公网安备 11010802027423号
京公网安备 11010802027423号