Cell Reports ( IF 7.5 ) Pub Date : 2021-01-19 , DOI: 10.1016/j.celrep.2020.108657 James Longden 1 , Xavier Robin 1 , Mathias Engel 2 , Jesper Ferkinghoff-Borg 1 , Ida Kjær 3 , Ivan D Horak 3 , Mikkel W Pedersen 3 , Rune Linding 4
|
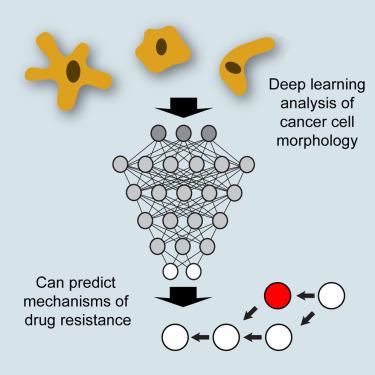
It is well known that the development of drug resistance in cancer cells can lead to changes in cell morphology. Here, we describe the use of deep neural networks to analyze this relationship, demonstrating that complex cell morphologies can encode states of signaling networks and unravel cellular mechanisms hidden to conventional approaches. We perform high-content screening of 17 cancer cell lines, generating more than 500 billion data points from ∼850 million cells. We analyze these data using a deep learning model, resulting in the identification of a continuous 27-dimension space describing all of the observed cell morphologies. From its morphology alone, we could thus predict whether a cell was resistant to ErbB-family drugs, with an accuracy of 74%, and predict the potential mechanism of resistance, subsequently validating the role of MET and insulin-like growth factor 1 receptor (IGF1R) as drivers of cetuximab resistance in in vitro models of lung and head/neck cancer.
中文翻译:

深度神经网络从连续的细胞形态空间中识别 ErbB 家族耐药性的信号机制
众所周知,癌细胞中耐药性的发展会导致细胞形态发生变化。在这里,我们描述了使用深度神经网络来分析这种关系,证明复杂的细胞形态可以编码信号网络的状态并解开隐藏在传统方法中的细胞机制。我们对 17 种癌细胞系进行高内涵筛选,从约 8.5 亿个细胞中生成超过 5000 亿个数据点。我们使用深度学习模型分析这些数据,从而识别出描述所有观察到的细胞形态的连续 27 维空间。因此,仅从其形态学来看,我们就可以预测细胞是否对 ErbB 家族药物产生耐药性,准确率为 74%,并预测耐药的潜在机制,肺癌和头颈癌的体外模型。











































 京公网安备 11010802027423号
京公网安备 11010802027423号