当前位置:
X-MOL 学术
›
J. Mol. Recognit.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
SNB‐PSSM: A spatial neighbor‐based PSSM used for protein–RNA binding site prediction
Journal of Molecular Recognition ( IF 2.3 ) Pub Date : 2021-01-14 , DOI: 10.1002/jmr.2887 Yang Liu 1 , Weikang Gong 1 , Zhen Yang 1 , Chunhua Li 1
Journal of Molecular Recognition ( IF 2.3 ) Pub Date : 2021-01-14 , DOI: 10.1002/jmr.2887 Yang Liu 1 , Weikang Gong 1 , Zhen Yang 1 , Chunhua Li 1
Affiliation
|
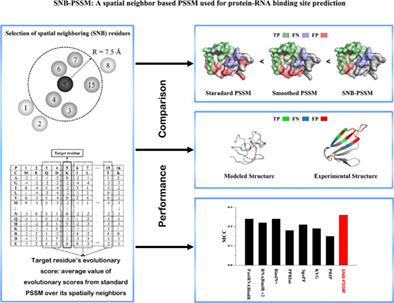
Protein–RNA interactions play essential roles in a wide variety of biological processes. Recognition of RNA‐binding residues on proteins has been a challenging problem. Most of methods utilize the position‐specific scoring matrix (PSSM). It has been found that considering the evolutionary information of sequence neighboring residues can improve the prediction. In this work, we introduce a novel method SNB‐PSSM (spatial neighbor–based PSSM) combined with the structure window scheme where the evolutionary information of spatially neighboring residues is considered. The results show our method consistently outperforms the standard and smoothed PSSM methods. Tested on multiple datasets, this approach shows an encouraging performance compared with RNABindRPlus, BindN+, PPRInt, xypan, Predict_RBP, SpaPF, PRNA, and KYG, although is inferior to RNAProSite, RBscore, and aaRNA. In addition, since our method is not sensitive to protein structure changes, it can be applied well on binding site predictions of modeled structures. Thus, the result also suggests the evolution of binding sites is spatially cooperative. The proposed method as an effective tool of considering evolutionary information can be widely used for the nucleic acid–/protein‐binding site prediction and functional motif finding.
中文翻译:

SNB-PSSM:用于蛋白质-RNA结合位点预测的基于空间邻居的PSSM
蛋白质-RNA 相互作用在多种生物过程中发挥着重要作用。识别蛋白质上的 RNA 结合残基一直是一个具有挑战性的问题。大多数方法使用特定位置评分矩阵(PSSM)。已经发现,考虑序列相邻残基的进化信息可以改进预测。在这项工作中,我们介绍了一种新的方法 SNB-PSSM(基于空间邻居的 PSSM),结合结构窗口方案,其中考虑了空间相邻残基的进化信息。结果表明,我们的方法始终优于标准和平滑 PSSM 方法。在多个数据集上进行测试,与 RNABindRPlus、BindN+、PPRInt、xypan、Predict_RBP、SpaPF、PRNA 和 KYG 相比,这种方法显示出令人鼓舞的性能,尽管不如 RNAProSite,RBscore 和 aaRNA。此外,由于我们的方法对蛋白质结构变化不敏感,因此可以很好地应用于模型结构的结合位点预测。因此,结果还表明结合位点的进化是空间合作的。所提出的方法作为考虑进化信息的有效工具,可广泛用于核酸/蛋白质结合位点预测和功能基序发现。
更新日期:2021-01-14
中文翻译:

SNB-PSSM:用于蛋白质-RNA结合位点预测的基于空间邻居的PSSM
蛋白质-RNA 相互作用在多种生物过程中发挥着重要作用。识别蛋白质上的 RNA 结合残基一直是一个具有挑战性的问题。大多数方法使用特定位置评分矩阵(PSSM)。已经发现,考虑序列相邻残基的进化信息可以改进预测。在这项工作中,我们介绍了一种新的方法 SNB-PSSM(基于空间邻居的 PSSM),结合结构窗口方案,其中考虑了空间相邻残基的进化信息。结果表明,我们的方法始终优于标准和平滑 PSSM 方法。在多个数据集上进行测试,与 RNABindRPlus、BindN+、PPRInt、xypan、Predict_RBP、SpaPF、PRNA 和 KYG 相比,这种方法显示出令人鼓舞的性能,尽管不如 RNAProSite,RBscore 和 aaRNA。此外,由于我们的方法对蛋白质结构变化不敏感,因此可以很好地应用于模型结构的结合位点预测。因此,结果还表明结合位点的进化是空间合作的。所提出的方法作为考虑进化信息的有效工具,可广泛用于核酸/蛋白质结合位点预测和功能基序发现。











































 京公网安备 11010802027423号
京公网安备 11010802027423号