当前位置:
X-MOL 学术
›
Biotechnol. J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Oligonucleotide abundance biases aid design of a type IIS synthetic genomics framework with plant virome capacity
Biotechnology Journal ( IF 3.2 ) Pub Date : 2021-01-07 , DOI: 10.1002/biot.202000354 Fabio Pasin
Biotechnology Journal ( IF 3.2 ) Pub Date : 2021-01-07 , DOI: 10.1002/biot.202000354 Fabio Pasin
|
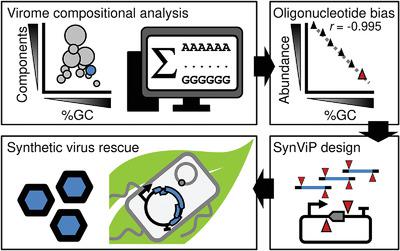
Synthetic genomics‐driven dematerialization of genetic resources facilitates flexible hypothesis testing and rapid product development. Biological sequences have compositional biases, which, I reasoned, could be exploited for engineering of enhanced synthetic genomics systems. In proof‐of‐concept assays reported herein, the abundance of random oligonucleotides in viral genomic components was analyzed and used for the rational design of a synthetic genomics framework with plant virome capacity (SynViP). Type IIS endonucleases with low abundance in the plant virome, as well as Golden Gate and No See'm principles were combined with DNA chemical synthesis for seamless viral clone assembly by one‐step digestion‐ligation. The framework described does not require subcloning steps, is insensitive to insert terminal sequences, and was used with linear and circular DNA molecules. Based on a digital template, DNA fragments were chemically synthesized and assembled by one‐step cloning to yield a scar‐free infectious clone of a plant virus suitable for Agrobacterium‐mediated delivery. SynViP allowed rescue of a genuine virus without biological material, and has the potential to greatly accelerate biological characterization and engineering of plant viruses as well as derived biotechnological tools. Finally, computational identification of compositional biases in biological sequences might become a common standard to aid scalable biosystems design and engineering.
中文翻译:

寡核苷酸丰度偏差可帮助设计具有植物病毒能力的IIS型合成基因组学框架
合成基因组学驱动的遗传资源去物质化有助于灵活的假设检验和快速的产品开发。我认为,生物学序列具有成分偏倚,可以用于增强合成基因组学系统的工程设计。在本文报道的概念验证试验中,分析了病毒基因组组件中大量随机寡核苷酸,并将其用于具有植物病毒能力(SynViP)的合成基因组框架的合理设计。植物病毒中低丰度的IIS型IIS核酸内切酶以及Golden Gate和No See'm原理与DNA化学合成相结合,可通过一步消化连接实现无缝病毒克隆装配。所描述的框架不需要亚克隆步骤,对插入末端序列不敏感,并与线性和环状DNA分子一起使用。根据数字模板,化学合成DNA片段,并通过一步克隆进行组装,以产生适用于植物病毒的无疤痕传染性克隆。农杆菌介导的传递。SynViP可以在没有生物物质的情况下拯救真正的病毒,并具有极大地加速植物病毒以及衍生生物技术工具的生物学表征和工程改造的潜力。最后,生物序列中组成偏差的计算鉴定可能会成为帮助可扩展生物系统设计和工程化的通用标准。
更新日期:2021-01-07
中文翻译:

寡核苷酸丰度偏差可帮助设计具有植物病毒能力的IIS型合成基因组学框架
合成基因组学驱动的遗传资源去物质化有助于灵活的假设检验和快速的产品开发。我认为,生物学序列具有成分偏倚,可以用于增强合成基因组学系统的工程设计。在本文报道的概念验证试验中,分析了病毒基因组组件中大量随机寡核苷酸,并将其用于具有植物病毒能力(SynViP)的合成基因组框架的合理设计。植物病毒中低丰度的IIS型IIS核酸内切酶以及Golden Gate和No See'm原理与DNA化学合成相结合,可通过一步消化连接实现无缝病毒克隆装配。所描述的框架不需要亚克隆步骤,对插入末端序列不敏感,并与线性和环状DNA分子一起使用。根据数字模板,化学合成DNA片段,并通过一步克隆进行组装,以产生适用于植物病毒的无疤痕传染性克隆。农杆菌介导的传递。SynViP可以在没有生物物质的情况下拯救真正的病毒,并具有极大地加速植物病毒以及衍生生物技术工具的生物学表征和工程改造的潜力。最后,生物序列中组成偏差的计算鉴定可能会成为帮助可扩展生物系统设计和工程化的通用标准。











































 京公网安备 11010802027423号
京公网安备 11010802027423号