Current Biology ( IF 9.2 ) Pub Date : 2021-01-07 , DOI: 10.1016/j.cub.2020.12.018 Gloria X Reyes 1 , Anna Kolodziejczak 2 , Lovely Jael Paul Solomon Devakumar 3 , Takashi Kubota 3 , Richard D Kolodner 4 , Christopher D Putnam 5 , Hans Hombauer 6
|
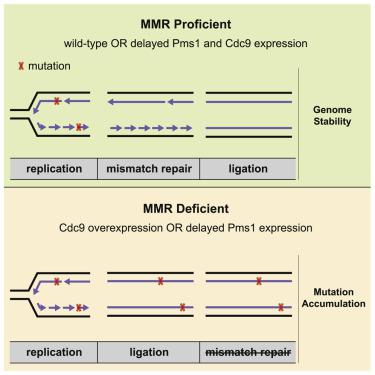
Mismatch repair (MMR) safeguards genome stability through recognition and excision of DNA replication errors.1, 2, 3, 4 How eukaryotic MMR targets the newly replicated strand in vivo has not been established. MMR reactions reconstituted in vitro are directed to the strand containing a preexisting nick or gap,5, 6, 7, 8 suggesting that strand discontinuities could act as discrimination signals. Another candidate is the proliferating cell nuclear antigen (PCNA) that is loaded at replication forks and is required for the activation of Mlh1-Pms1 endonuclease.7, 8, 9 Here, we discovered that overexpression of DNA ligase I (Cdc9) in Saccharomyces cerevisiae causes elevated mutation rates and increased chromatin-bound PCNA levels and accumulation of Pms1 foci that are MMR intermediates, suggesting that premature ligation of replication-associated nicks interferes with MMR. We showed that yeast Pms1 expression is mainly restricted to S phase, in agreement with the temporal coupling between MMR and DNA replication.10 Restricting Pms1 expression to the G2/M phase caused a mutator phenotype that was exacerbated in the absence of the exonuclease Exo1. This mutator phenotype was largely suppressed by increasing the lifetime of replication-associated DNA nicks, either by reducing or delaying Cdc9 ligase activity in vivo. Therefore, Cdc9 dictates a window of time for MMR determined by transient DNA nicks that direct the Mlh1-Pms1 in a strand-specific manner. Because DNA nicks occur on both newly synthesized leading and lagging strands,11 these results establish a general mechanism for targeting MMR to the newly synthesized DNA, thus preventing the accumulation of mutations that underlie the development of human cancer.
中文翻译:

新复制的 DNA 的连接控制 DNA 错配修复的时间
错配修复 (MMR) 通过识别和切除 DNA 复制错误来保护基因组稳定性。1, 2, 3, 4 真核生物 MMR 如何在体内靶向新复制的链尚未确定。体外重组的 MMR 反应针对包含预先存在的缺口或间隙的链 5、6、7、8,这表明链的不连续性可以作为识别信号。另一个候选者是增殖细胞核抗原 (PCNA),它装载在复制叉上,是激活 Mlh1-Pms1 核酸内切酶所必需的。7, 8, 9 在这里,我们发现酿酒酵母中 DNA 连接酶 I (Cdc9) 的过表达导致突变率升高和染色质结合 PCNA 水平增加,以及作为 MMR 中间体的 Pms1 病灶的积累,表明复制相关切口的过早连接会干扰 MMR。我们发现酵母 Pms1 表达主要局限于 S 期,这与 MMR 和 DNA 复制之间的时间耦合一致。10将 Pms1 表达限制在 G2/M 期会导致突变表型在没有外切核酸酶 Exo1 的情况下加剧。通过增加复制相关 DNA 切口的寿命,通过减少或延迟体内Cdc9 连接酶活性,这种突变表型在很大程度上受到抑制. 因此,Cdc9 规定了 MMR 的时间窗口,该时间窗口由以链特异性方式引导 Mlh1-Pms1 的瞬时 DNA 切口确定。由于新合成的前导链和滞后链上都会出现 DNA 切口,11这些结果确立了将 MMR 靶向新合成的 DNA 的一般机制,从而防止突变的积累,这些突变是人类癌症发展的基础。



























 京公网安备 11010802027423号
京公网安备 11010802027423号