当前位置:
X-MOL 学术
›
Ann. Appl. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Optimising accuracy of performance predictions using available morphophysiological information in wheat breeding germplasm
Annals of Applied Biology ( IF 2.2 ) Pub Date : 2021-01-04 , DOI: 10.1111/aab.12672 Sunčica Guberac 1 , Vlatko Galić 2 , Andrijana Rebekić 1 , Tihomir Čupić 2 , Sonja Petrović 1
Annals of Applied Biology ( IF 2.2 ) Pub Date : 2021-01-04 , DOI: 10.1111/aab.12672 Sunčica Guberac 1 , Vlatko Galić 2 , Andrijana Rebekić 1 , Tihomir Čupić 2 , Sonja Petrović 1
Affiliation
|
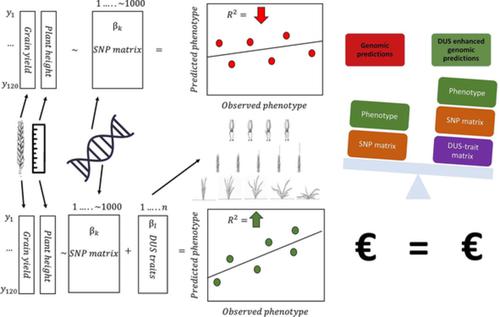
Wheat breeding programmes often harbour large number of developed progenies. Testing of all progenies in many target environments is not considered cost‐effective, so the genomic predictions are employed. Genomic predictions might be enhanced with adaptational traits given the cost‐effectiveness of their phenotyping. The aim of this study was to assess effectiveness of adding a priori available phenotyping data of adaptational morpho‐physiological traits to genomic predictions in Bayesian framework. The panel of 120 winter wheat genotypes was sown over four growing seasons (2013/2014–2016/2017) in field conditions in two replicates and phenotyped for target traits grain yield and plant height along with nine distinctness, uniformity and stability (DUS) traits. Five genotype groups were defined by the K‐means clustering method based on nine DUS traits. DUS traits were used as fixed covariates in five different genomic prediction models with different assumptions about the distribution densities of marker effects: Bayes A, Bayes B, Bayes C, Bayesian ridge regression and Bayesian lasso. Adding DUS traits as covariates significantly improved accuracies and reduced the root mean square errors (RMSEP) of genomic predictions. Marginal differences between different models were observed. Adding covariates to genomic prediction models might be good strategy to improve accuracies of the predictions by accounting for environmental adaptations, provided their a priori availability or low costs of additional phenotyping.
中文翻译:

利用可获得的形态生理信息优化小麦育种种质表现预测的准确性
小麦育种计划通常包含大量的后代。在许多目标环境中测试所有后代并不具有成本效益,因此采用了基因组预测。鉴于表型的成本效益,适应性状可能会增强基因组预测。本研究的目的是评估在贝叶斯框架的基因组预测中添加适应性形态生理特征的先验可用表型数据的有效性。在四个生长季节(2013 / 2014–2016 / 2017)上,在田间条件下播种了120种冬小麦基因型,进行了两次重复试验,并根据目标性状,单产和株高以及九种显着性,均匀性和稳定性(DUS)性状进行了表型分析。 。通过基于9种DUS性状的K-均值聚类方法定义了5个基因型组。DUS性状在五个不同的基因组预测模型中用作固定协变量,对标记效应的分布密度有不同的假设:贝叶斯A,贝叶斯B,贝叶斯C,贝叶斯岭回归和贝叶斯套索。将DUS性状添加为协变量可显着提高准确性,并减少基因组预测的均方根误差(RMSEP)。观察到不同模型之间的边际差异。将协变量添加到基因组预测模型中可能是通过考虑环境适应性来提高预测准确性的好策略,前提是它们具有先验可用性或附加表型的低成本。DUS性状在五个不同的基因组预测模型中用作固定协变量,对标记效应的分布密度有不同的假设:贝叶斯A,贝叶斯B,贝叶斯C,贝叶斯岭回归和贝叶斯套索。将DUS性状添加为协变量可显着提高准确性,并减少基因组预测的均方根误差(RMSEP)。观察到不同模型之间的边际差异。将协变量添加到基因组预测模型中可能是通过考虑环境适应性来提高预测准确性的好策略,前提是它们具有先验可用性或附加表型的低成本。DUS性状在五个不同的基因组预测模型中用作固定协变量,对标记效应的分布密度有不同的假设:贝叶斯A,贝叶斯B,贝叶斯C,贝叶斯岭回归和贝叶斯套索。将DUS性状添加为协变量可显着提高准确性,并减少基因组预测的均方根误差(RMSEP)。观察到不同模型之间的边际差异。将协变量添加到基因组预测模型中可能是通过考虑环境适应性来提高预测准确性的好策略,前提是它们具有先验可用性或附加表型的低成本。将DUS性状添加为协变量可显着提高准确性,并减少基因组预测的均方根误差(RMSEP)。观察到不同模型之间的边际差异。将协变量添加到基因组预测模型中可能是通过考虑环境适应性来提高预测准确性的好策略,前提是它们具有先验可用性或附加表型的低成本。将DUS性状添加为协变量可显着提高准确性,并减少基因组预测的均方根误差(RMSEP)。观察到不同模型之间的边际差异。将协变量添加到基因组预测模型中可能是通过考虑环境适应性来提高预测准确性的好策略,前提是它们具有先验可用性或附加表型的低成本。
更新日期:2021-01-04
中文翻译:

利用可获得的形态生理信息优化小麦育种种质表现预测的准确性
小麦育种计划通常包含大量的后代。在许多目标环境中测试所有后代并不具有成本效益,因此采用了基因组预测。鉴于表型的成本效益,适应性状可能会增强基因组预测。本研究的目的是评估在贝叶斯框架的基因组预测中添加适应性形态生理特征的先验可用表型数据的有效性。在四个生长季节(2013 / 2014–2016 / 2017)上,在田间条件下播种了120种冬小麦基因型,进行了两次重复试验,并根据目标性状,单产和株高以及九种显着性,均匀性和稳定性(DUS)性状进行了表型分析。 。通过基于9种DUS性状的K-均值聚类方法定义了5个基因型组。DUS性状在五个不同的基因组预测模型中用作固定协变量,对标记效应的分布密度有不同的假设:贝叶斯A,贝叶斯B,贝叶斯C,贝叶斯岭回归和贝叶斯套索。将DUS性状添加为协变量可显着提高准确性,并减少基因组预测的均方根误差(RMSEP)。观察到不同模型之间的边际差异。将协变量添加到基因组预测模型中可能是通过考虑环境适应性来提高预测准确性的好策略,前提是它们具有先验可用性或附加表型的低成本。DUS性状在五个不同的基因组预测模型中用作固定协变量,对标记效应的分布密度有不同的假设:贝叶斯A,贝叶斯B,贝叶斯C,贝叶斯岭回归和贝叶斯套索。将DUS性状添加为协变量可显着提高准确性,并减少基因组预测的均方根误差(RMSEP)。观察到不同模型之间的边际差异。将协变量添加到基因组预测模型中可能是通过考虑环境适应性来提高预测准确性的好策略,前提是它们具有先验可用性或附加表型的低成本。DUS性状在五个不同的基因组预测模型中用作固定协变量,对标记效应的分布密度有不同的假设:贝叶斯A,贝叶斯B,贝叶斯C,贝叶斯岭回归和贝叶斯套索。将DUS性状添加为协变量可显着提高准确性,并减少基因组预测的均方根误差(RMSEP)。观察到不同模型之间的边际差异。将协变量添加到基因组预测模型中可能是通过考虑环境适应性来提高预测准确性的好策略,前提是它们具有先验可用性或附加表型的低成本。将DUS性状添加为协变量可显着提高准确性,并减少基因组预测的均方根误差(RMSEP)。观察到不同模型之间的边际差异。将协变量添加到基因组预测模型中可能是通过考虑环境适应性来提高预测准确性的好策略,前提是它们具有先验可用性或附加表型的低成本。将DUS性状添加为协变量可显着提高准确性,并减少基因组预测的均方根误差(RMSEP)。观察到不同模型之间的边际差异。将协变量添加到基因组预测模型中可能是通过考虑环境适应性来提高预测准确性的好策略,前提是它们具有先验可用性或附加表型的低成本。











































 京公网安备 11010802027423号
京公网安备 11010802027423号