当前位置:
X-MOL 学术
›
Biotechnol. Bioeng.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Incorporating RNA‐Seq transcriptomics into glycosylation‐integrating metabolic network modelling kinetics: Multiomic Chinese hamster ovary (CHO) cell bioreactors
Biotechnology and Bioengineering ( IF 3.5 ) Pub Date : 2021-01-05 , DOI: 10.1002/bit.27660 Lara Bezjak 1 , Vivian Erklavec Zajec 1 , Špela Baebler 2 , Tjaša Stare 2 , Kristina Gruden 2 , Andrej Pohar 1 , Uroš Novak 1 , Blaž Likozar 1
Biotechnology and Bioengineering ( IF 3.5 ) Pub Date : 2021-01-05 , DOI: 10.1002/bit.27660 Lara Bezjak 1 , Vivian Erklavec Zajec 1 , Špela Baebler 2 , Tjaša Stare 2 , Kristina Gruden 2 , Andrej Pohar 1 , Uroš Novak 1 , Blaž Likozar 1
Affiliation
|
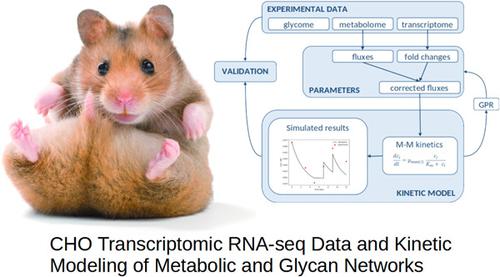
In this work, the kinetic model based on the previously developed metabolic and glycan reaction networks of the ovarian cells of the Chinese hamster ovary (CHO) cell line was improved by the inclusion of transcriptomic data that took into account the values of the RPKM gene (Reads per Kilobase of Exon per Million Reads Mapped). The transcriptomic (RNASeq) data were obtained together with metabolic and glycan data from the literature, and the concentrations with RPKM values were collected at several points in time from two fed‐batch processes. First, the fluxes were determined by regression analysis of the metabolic data, then these fluxes were corrected by using the fold change in gene expression as a measure of enzyme concentrations. Next, the corrected fluxes in the kinetic model were used to calculate the concentration profiles of the metabolites, and literature data were used to evaluate the predicted results of the model. Compared to other studies where the concentration profiles of CHO cell metabolites were described using a kinetic model without consideration of RNA‐Seq data to correct the fluxes, this model is unique. The additional integration of transcriptomic data led to better predictions of metabolic concentrations in the fed‐batch process, which is a significant improvement of the modelling technique used.
更新日期:2021-01-05











































 京公网安备 11010802027423号
京公网安备 11010802027423号