当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Functions of Essential Genes and a Scale-Free Protein Interaction Network Revealed by Structure-Based Function and Interaction Prediction for a Minimal Genome
Journal of Proteome Research ( IF 4.4 ) Pub Date : 2021-01-04 , DOI: 10.1021/acs.jproteome.0c00359 Chengxin Zhang 1 , Wei Zheng 1 , Micah Cheng 2 , Gilbert S Omenn 1, 3 , Peter L Freddolino 1, 4 , Yang Zhang 1, 4
Journal of Proteome Research ( IF 4.4 ) Pub Date : 2021-01-04 , DOI: 10.1021/acs.jproteome.0c00359 Chengxin Zhang 1 , Wei Zheng 1 , Micah Cheng 2 , Gilbert S Omenn 1, 3 , Peter L Freddolino 1, 4 , Yang Zhang 1, 4
Affiliation
|
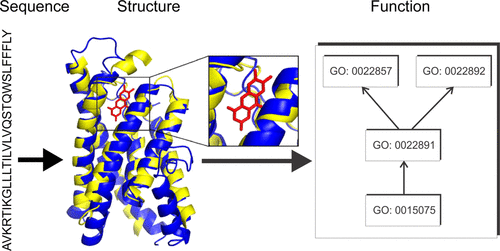
When the JCVI-syn3.0 genome was designed and implemented in 2016 as the minimal genome of a free-living organism, approximately one-third of the 438 protein-coding genes had no known function. Subsequent refinement into JCVI-syn3A led to inclusion of 16 additional protein-coding genes, including several unknown functions, resulting in an improved growth phenotype. Here, we seek to unveil the biological roles and protein–protein interaction (PPI) networks for these poorly characterized proteins using state-of-the-art deep learning contact-assisted structure prediction, followed by structure-based annotation of functions and PPI predictions. Our pipeline is able to confidently assign functions for many previously unannotated proteins such as putative vitamin transporters, which suggest the importance of nutrient uptake even in a minimized genome. Remarkably, despite the artificial selection of genes in the minimal syn3 genome, our reconstructed PPI network still shows a power law distribution of node degrees typical of naturally evolved bacterial PPI networks. Making use of our framework for combined structure/function/interaction modeling, we are able to identify both fundamental aspects of network biology that are retained in a minimal proteome and additional essential functions not yet recognized among the poorly annotated components of the syn3.0 and syn3A proteomes.
中文翻译:

最小基因组基于结构的功能和相互作用预测揭示了必需基因的功能和无标度蛋白质相互作用网络
当 JCVI-syn3.0 基因组于 2016 年设计并实现为自由生物体的最小基因组时,438 个蛋白质编码基因中约有三分之一没有已知的功能。随后对 JCVI-syn3A 进行改进,纳入了 16 个额外的蛋白质编码基因,其中包括几个未知的功能,从而改善了生长表型。在这里,我们试图使用最先进的深度学习接触辅助结构预测来揭示这些特征较差的蛋白质的生物学作用和蛋白质-蛋白质相互作用 (PPI) 网络,然后进行基于结构的功能注释和 PPI 预测。我们的管道能够自信地为许多以前未注释的蛋白质(例如假定的维生素转运蛋白)分配功能,这表明即使在最小化的基因组中营养吸收的重要性。值得注意的是,尽管在最小 syn3 基因组中对基因进行了人工选择,但我们重建的 PPI 网络仍然显示出自然进化的细菌 PPI 网络典型的节点度幂律分布。利用我们的组合结构/功能/相互作用建模框架,我们能够识别保留在最小蛋白质组中的网络生物学的基本方面,以及在 syn3.0 注释不良的组件中尚未识别的其他基本功能。 syn3A 蛋白质组。
更新日期:2021-02-05
中文翻译:

最小基因组基于结构的功能和相互作用预测揭示了必需基因的功能和无标度蛋白质相互作用网络
当 JCVI-syn3.0 基因组于 2016 年设计并实现为自由生物体的最小基因组时,438 个蛋白质编码基因中约有三分之一没有已知的功能。随后对 JCVI-syn3A 进行改进,纳入了 16 个额外的蛋白质编码基因,其中包括几个未知的功能,从而改善了生长表型。在这里,我们试图使用最先进的深度学习接触辅助结构预测来揭示这些特征较差的蛋白质的生物学作用和蛋白质-蛋白质相互作用 (PPI) 网络,然后进行基于结构的功能注释和 PPI 预测。我们的管道能够自信地为许多以前未注释的蛋白质(例如假定的维生素转运蛋白)分配功能,这表明即使在最小化的基因组中营养吸收的重要性。值得注意的是,尽管在最小 syn3 基因组中对基因进行了人工选择,但我们重建的 PPI 网络仍然显示出自然进化的细菌 PPI 网络典型的节点度幂律分布。利用我们的组合结构/功能/相互作用建模框架,我们能够识别保留在最小蛋白质组中的网络生物学的基本方面,以及在 syn3.0 注释不良的组件中尚未识别的其他基本功能。 syn3A 蛋白质组。



























 京公网安备 11010802027423号
京公网安备 11010802027423号