当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Metaproteomics Analysis of SARS-CoV-2-Infected Patient Samples Reveals Presence of Potential Coinfecting Microorganisms
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2021-01-04 , DOI: 10.1021/acs.jproteome.0c00822 Peter S Thuy-Boun 1 , Subina Mehta 2 , Bjoern Gruening 3 , Thomas McGowan 2 , An Nguyen 2 , Andrew T Rajczewski 2 , James E Johnson 2 , Timothy J Griffin 2 , Dennis W Wolan 1 , Pratik D Jagtap 2
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2021-01-04 , DOI: 10.1021/acs.jproteome.0c00822 Peter S Thuy-Boun 1 , Subina Mehta 2 , Bjoern Gruening 3 , Thomas McGowan 2 , An Nguyen 2 , Andrew T Rajczewski 2 , James E Johnson 2 , Timothy J Griffin 2 , Dennis W Wolan 1 , Pratik D Jagtap 2
Affiliation
|
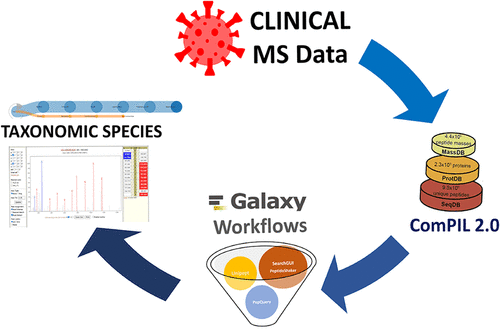
In this Letter, we reanalyze published mass spectrometry data sets of clinical samples with a focus on determining the coinfection status of individuals infected with SARS-CoV-2 coronavirus. We demonstrate the use of ComPIL 2.0 software along with a metaproteomics workflow within the Galaxy platform to detect cohabitating potential pathogens in COVID-19 patients using mass spectrometry-based analysis. From a sample collected from gargling solutions, we detected Streptococcus pneumoniae (opportunistic and multidrug-resistant pathogen) and Lactobacillus rhamnosus (a probiotic component) along with SARS-Cov-2. We could also detect Pseudomonas sps. Bc-h from COVID-19 positive samples and Acinetobacter ursingii and Pseudomonas monteilii from COVID-19 negative samples collected from oro- and nasopharyngeal samples. We believe that the early detection and characterization of coinfections by using metaproteomics from COVID-19 patients will potentially impact the diagnosis and treatment of patients affected by SARS-CoV-2 infection.
更新日期:2021-02-05











































 京公网安备 11010802027423号
京公网安备 11010802027423号