当前位置:
X-MOL 学术
›
J. Mol. Recognit.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Exploring insights of syntaxin superfamily proteins from Entamoeba histolytica: a prospective simulation, protein‐protein interaction, and docking study
Journal of Molecular Recognition ( IF 2.3 ) Pub Date : 2021-01-03 , DOI: 10.1002/jmr.2886 Sagar Batra 1 , Puranjaya Pancholi 1 , Mrinalini Roy 1 , Sanket Kaushik 1 , Anupam Jyoti 1 , Kuldeep Verma 2 , Vijay Kumar Srivastava 1
Journal of Molecular Recognition ( IF 2.3 ) Pub Date : 2021-01-03 , DOI: 10.1002/jmr.2886 Sagar Batra 1 , Puranjaya Pancholi 1 , Mrinalini Roy 1 , Sanket Kaushik 1 , Anupam Jyoti 1 , Kuldeep Verma 2 , Vijay Kumar Srivastava 1
Affiliation
|
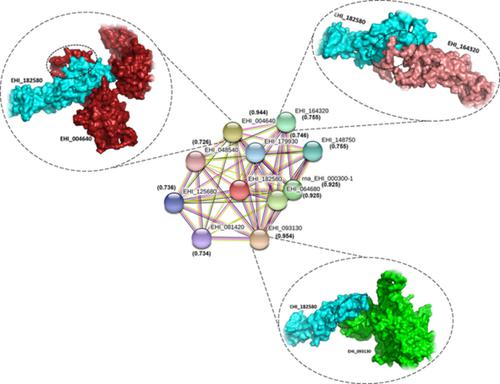
Entamoeba histolytica (Eh), a parasitic protozoan and the causative agent of invasive Amoebiasis, invade the host tissue through an effective secretory pathway. There are several lines of evidence suggesting that amoebic trophozoite pore‐forming complex amoebapore and a large class of proteases enzymes including rhomboid proteases, cysteine proteases, and metalloproteases are implicated in host tissue invasion. For successful delivery of these molecules/cargos, trophozoites heavily rely on sorting machinery from the endoplasmic reticulum, Golgi to plasma membrane. Although, sole secretion machinery in E. histolytica is not characterized yet. Therefore, here our aim is to understand the properties of key molecules N‐ethylmaleimide‐sensitive fusion protein attached to protein receptors (SNAREs) in E. histolytica. SNAREs proteins are an important component of the membrane‐trafficking machinery and have been associated in a range of processes including vesicle tethering, fusion as well as specificity of vesicular transport in all eukaryotic cells. SNARE proteins are architecturally simple, categorized by the presence of one copy of a homologous coiled‐coil forming motif. However, the structural information and protein‐protein interaction study of Eh‐associated syntaxin proteins are still not known. Here, we characterize the syntaxin 1 like molecule and VAMP from Eh through physiochemical profiling, modeling, atomistic simulation, protein‐protein interaction, and docking approaches on the proteins containing SNARE and synaptobrevin domain. The modeled structures and the critical residues recognized through protein interaction and docking study may provide better structural and functional insights into these proteins and may aid in the development of newer diagnostic assays.
中文翻译:

探索溶组织内阿米巴突触蛋白超家族蛋白的见解:前瞻性模拟、蛋白质-蛋白质相互作用和对接研究
溶组织内阿米巴( Eh ) 是一种寄生原生动物和侵袭性阿米巴病的病原体,通过有效的分泌途径侵入宿主组织。有几条证据表明,阿米巴滋养体成孔复合物阿米巴孔和一大类蛋白酶,包括菱形蛋白酶、半胱氨酸蛋白酶和金属蛋白酶,与宿主组织的侵袭有关。为了成功运送这些分子/货物,滋养体严重依赖从内质网、高尔基体到质膜的分拣机械。虽然,溶组织大肠杆菌中的唯一分泌机制尚未确定。因此,我们的目标是了解关键分子N的性质与溶组织大肠杆菌中的蛋白质受体 (SNARE) 连接的-乙基马来酰亚胺敏感性融合蛋白。SNAREs 蛋白是膜运输机制的重要组成部分,并与一系列过程相关,包括囊泡束缚、融合以及所有真核细胞中囊泡转运的特异性。SNARE 蛋白在结构上很简单,通过存在一个同源卷曲螺旋形成基序的拷贝来分类。然而,Eh相关突触蛋白的结构信息和蛋白质-蛋白质相互作用研究仍然未知。在这里,我们描述了来自Eh的突触蛋白 1 样分子和 VAMP通过对含有 SNARE 和 synaptobrevin 结构域的蛋白质进行物理化学分析、建模、原子模拟、蛋白质-蛋白质相互作用和对接方法。通过蛋白质相互作用和对接研究识别的建模结构和关键残基可以为这些蛋白质提供更好的结构和功能见解,并可能有助于开发新的诊断分析。
更新日期:2021-01-03
中文翻译:

探索溶组织内阿米巴突触蛋白超家族蛋白的见解:前瞻性模拟、蛋白质-蛋白质相互作用和对接研究
溶组织内阿米巴( Eh ) 是一种寄生原生动物和侵袭性阿米巴病的病原体,通过有效的分泌途径侵入宿主组织。有几条证据表明,阿米巴滋养体成孔复合物阿米巴孔和一大类蛋白酶,包括菱形蛋白酶、半胱氨酸蛋白酶和金属蛋白酶,与宿主组织的侵袭有关。为了成功运送这些分子/货物,滋养体严重依赖从内质网、高尔基体到质膜的分拣机械。虽然,溶组织大肠杆菌中的唯一分泌机制尚未确定。因此,我们的目标是了解关键分子N的性质与溶组织大肠杆菌中的蛋白质受体 (SNARE) 连接的-乙基马来酰亚胺敏感性融合蛋白。SNAREs 蛋白是膜运输机制的重要组成部分,并与一系列过程相关,包括囊泡束缚、融合以及所有真核细胞中囊泡转运的特异性。SNARE 蛋白在结构上很简单,通过存在一个同源卷曲螺旋形成基序的拷贝来分类。然而,Eh相关突触蛋白的结构信息和蛋白质-蛋白质相互作用研究仍然未知。在这里,我们描述了来自Eh的突触蛋白 1 样分子和 VAMP通过对含有 SNARE 和 synaptobrevin 结构域的蛋白质进行物理化学分析、建模、原子模拟、蛋白质-蛋白质相互作用和对接方法。通过蛋白质相互作用和对接研究识别的建模结构和关键残基可以为这些蛋白质提供更好的结构和功能见解,并可能有助于开发新的诊断分析。











































 京公网安备 11010802027423号
京公网安备 11010802027423号