当前位置:
X-MOL 学术
›
Biotechnol. J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A deep learning approach to evaluate the feasibility of enzymatic reactions generated by retrobiosynthesis
Biotechnology Journal ( IF 3.2 ) Pub Date : 2021-01-02 , DOI: 10.1002/biot.202000605 Yeji Kim 1, 2, 3 , Jae Yong Ryu 4 , Hyun Uk Kim 2, 3, 5 , Woo Dae Jang 1, 2, 3 , Sang Yup Lee 1, 2, 3
Biotechnology Journal ( IF 3.2 ) Pub Date : 2021-01-02 , DOI: 10.1002/biot.202000605 Yeji Kim 1, 2, 3 , Jae Yong Ryu 4 , Hyun Uk Kim 2, 3, 5 , Woo Dae Jang 1, 2, 3 , Sang Yup Lee 1, 2, 3
Affiliation
|
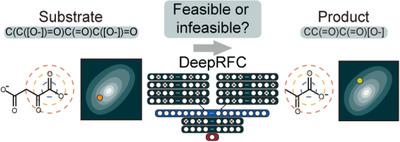
Retrobiosynthesis allows the designing of novel biosynthetic pathways for the production of chemicals and materials through metabolic engineering, but generates a large number of reactions beyond the experimental feasibility. Thus, an effective method that can reduce a large number of the initially predicted enzymatic reactions has been needed. Here, we present Deep learning‐based Reaction Feasibility Checker (DeepRFC) to classify the feasibility of a given enzymatic reaction with high performance and speed. DeepRFC is designed to receive Simplified Molecular‐Input Line‐Entry System (SMILES) strings of a reactant pair, which is defined as a substrate and a product of a reaction, as an input, and evaluates whether the input reaction is feasible. A deep neural network is selected for DeepRFC as it leads to better classification performance than five other representative machine learning methods examined. For validation, the performance of DeepRFC is compared with another in‐house reaction feasibility checker that uses the concept of reaction similarity. Finally, the use of DeepRFC is demonstrated for the retrobiosynthesis‐based design of novel one‐carbon assimilation pathways. DeepRFC will allow retrobiosynthesis to be more practical for metabolic engineering applications by efficiently screening a large number of retrobiosynthesis‐derived enzymatic reactions. DeepRFC is freely available at https://bitbucket.org/kaistsystemsbiology/deeprfc.
中文翻译:

评估逆向生物合成产生的酶促反应可行性的深度学习方法
逆向生物合成允许设计用于通过代谢工程生产化学品和材料的新型生物合成途径,但会产生超出实验可行性的大量反应。因此,需要一种可以减少大量最初预测的酶促反应的有效方法。在这里,我们介绍基于深度学习的反应可行性检查器(DeepRFC),以高性能和高速度对给定酶促反应的可行性进行分类。DeepRFC旨在接收被定义为底物和反应产物的反应物对的简化分子输入线进入系统(SMILES)字符串作为输入,并评估输入反应是否可行。为DeepRFC选择了深度神经网络,因为它比其他五种代表性的机器学习方法具有更好的分类性能。为了进行验证,将DeepRFC的性能与另一个使用反应相似性概念的内部反应可行性检查器进行比较。最后,证明了DeepRFC在新颖的一碳同化途径的基于逆向生物合成的设计中的使用。DeepRFC通过有效地筛选大量源自逆转录合成的酶促反应,使逆转录合成在代谢工程应用中更加实用。DeepRFC可从https://bitbucket.org/kaistsystemsbiology/deeprfc免费获得。将DeepRFC的性能与另一个使用反应相似性概念的内部反应可行性检查器进行比较。最后,证明了DeepRFC在新颖的一碳同化途径的基于逆向生物合成的设计中的使用。DeepRFC通过有效地筛选大量源自逆转录合成的酶促反应,可使逆转录合成在代谢工程应用中更加实用。DeepRFC可从https://bitbucket.org/kaistsystemsbiology/deeprfc免费获得。将DeepRFC的性能与另一个使用反应相似性概念的内部反应可行性检查器进行比较。最后,证明了DeepRFC在新颖的一碳同化途径的基于逆向生物合成的设计中的使用。DeepRFC通过有效地筛选大量源自逆转录合成的酶促反应,使逆转录合成在代谢工程应用中更加实用。DeepRFC可从https://bitbucket.org/kaistsystemsbiology/deeprfc免费获得。DeepRFC通过有效地筛选大量源自逆转录合成的酶促反应,使逆转录合成在代谢工程应用中更加实用。DeepRFC可从https://bitbucket.org/kaistsystemsbiology/deeprfc免费获得。DeepRFC通过有效地筛选大量源自逆转录合成的酶促反应,使逆转录合成在代谢工程应用中更加实用。DeepRFC可从https://bitbucket.org/kaistsystemsbiology/deeprfc免费获得。
更新日期:2021-01-02
中文翻译:

评估逆向生物合成产生的酶促反应可行性的深度学习方法
逆向生物合成允许设计用于通过代谢工程生产化学品和材料的新型生物合成途径,但会产生超出实验可行性的大量反应。因此,需要一种可以减少大量最初预测的酶促反应的有效方法。在这里,我们介绍基于深度学习的反应可行性检查器(DeepRFC),以高性能和高速度对给定酶促反应的可行性进行分类。DeepRFC旨在接收被定义为底物和反应产物的反应物对的简化分子输入线进入系统(SMILES)字符串作为输入,并评估输入反应是否可行。为DeepRFC选择了深度神经网络,因为它比其他五种代表性的机器学习方法具有更好的分类性能。为了进行验证,将DeepRFC的性能与另一个使用反应相似性概念的内部反应可行性检查器进行比较。最后,证明了DeepRFC在新颖的一碳同化途径的基于逆向生物合成的设计中的使用。DeepRFC通过有效地筛选大量源自逆转录合成的酶促反应,使逆转录合成在代谢工程应用中更加实用。DeepRFC可从https://bitbucket.org/kaistsystemsbiology/deeprfc免费获得。将DeepRFC的性能与另一个使用反应相似性概念的内部反应可行性检查器进行比较。最后,证明了DeepRFC在新颖的一碳同化途径的基于逆向生物合成的设计中的使用。DeepRFC通过有效地筛选大量源自逆转录合成的酶促反应,可使逆转录合成在代谢工程应用中更加实用。DeepRFC可从https://bitbucket.org/kaistsystemsbiology/deeprfc免费获得。将DeepRFC的性能与另一个使用反应相似性概念的内部反应可行性检查器进行比较。最后,证明了DeepRFC在新颖的一碳同化途径的基于逆向生物合成的设计中的使用。DeepRFC通过有效地筛选大量源自逆转录合成的酶促反应,使逆转录合成在代谢工程应用中更加实用。DeepRFC可从https://bitbucket.org/kaistsystemsbiology/deeprfc免费获得。DeepRFC通过有效地筛选大量源自逆转录合成的酶促反应,使逆转录合成在代谢工程应用中更加实用。DeepRFC可从https://bitbucket.org/kaistsystemsbiology/deeprfc免费获得。DeepRFC通过有效地筛选大量源自逆转录合成的酶促反应,使逆转录合成在代谢工程应用中更加实用。DeepRFC可从https://bitbucket.org/kaistsystemsbiology/deeprfc免费获得。











































 京公网安备 11010802027423号
京公网安备 11010802027423号