当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Full-Range Profiling of tRNA Modifications Using LC–MS/MS at Single-Base Resolution through a Site-Specific Cleavage Strategy
Analytical Chemistry ( IF 6.7 ) Pub Date : 2020-12-31 , DOI: 10.1021/acs.analchem.0c03307 Tong-Meng Yan 1 , Yu Pan 1 , Meng-Lan Yu 1 , Kua Hu 1 , Kai-Yue Cao 1 , Zhi-Hong Jiang 1
Analytical Chemistry ( IF 6.7 ) Pub Date : 2020-12-31 , DOI: 10.1021/acs.analchem.0c03307 Tong-Meng Yan 1 , Yu Pan 1 , Meng-Lan Yu 1 , Kua Hu 1 , Kai-Yue Cao 1 , Zhi-Hong Jiang 1
Affiliation
|
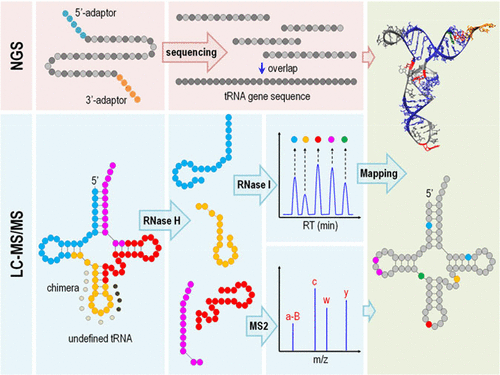
Transfer RNAs (tRNAs) are the most heavily modified RNA species. Liquid chromatography coupled with mass spectrometry (LC–MS/MS) is a powerful tool for characterizing tRNA modifications, which involves pretreating tRNAs with base-specific ribonucleases to produce smaller oligonucleotides amenable to MS. However, the quality and quantity of products from base-specific digestions are severely impacted by the base composition of tRNAs. This often leads to a loss of sequence information. Here, we report a method for the full-range profiling of tRNA modifications at single-base resolution by combining site-specific RNase H digestion with the LC–MS/MS and RNA-seq techniques. The key steps were designed to generate high-quality products of optimal lengths and ionization properties. A linear correlation between collision energies and the m/z of oligonucleotides significantly improved the information content of collision-induced dissociation (CID) spectra. False positives were eliminated by up to 95% using novel inclusion criteria for collecting a census of modifications. This method is illustrated by the mapping of mouse mitochondrial tRNAHis(GUG) and tRNAVal(UAC), which were hitherto not investigated. The identities and locations of the five species of modifications on these tRNAs were fully characterized. This approach is universally applicable to any tRNA species and provides an experimentally realizable pathway to the de novo sequencing of post-transcriptionally modified tRNAs with high sequence coverage.
中文翻译:

使用LC-MS / MS通过特定位点切割策略以单碱基分辨率对tRNA修饰进行全范围分析
转移RNA(tRNA)是修饰程度最高的RNA种类。液相色谱与质谱联用(LC-MS / MS)是表征tRNA修饰的强大工具,其中涉及使用碱基特异性核糖核酸对tRNA进行预处理,以产生适合MS的较小寡核苷酸。但是,tRNA的碱基组成会严重影响碱基特异性消化产物的质量和数量。这通常导致序列信息的丢失。在这里,我们报告了一种通过结合LC-MS / MS和RNA-seq技术结合位点特异性RNase H消化以单碱基分辨率对tRNA修饰进行全方位分析的方法。设计关键步骤是为了生成具有最佳长度和电离特性的高质量产品。碰撞能量与碰撞能量之间的线性关系m / z的寡核苷酸显着提高了碰撞诱导解离(CID)光谱的信息含量。使用新的纳入标准收集修改普查结果,误报率高达95%。迄今为止尚未研究的小鼠线粒体tRNA His(GUG)和tRNA Val(UAC)的作图说明了该方法。这些tRNA上的五个修饰物种的身份和位置已得到充分表征。这种方法普遍适用于任何tRNA种类,并提供了从实验上可实现的途径,以具有高序列覆盖率的转录后修饰tRNA从头测序。
更新日期:2021-01-26
中文翻译:

使用LC-MS / MS通过特定位点切割策略以单碱基分辨率对tRNA修饰进行全范围分析
转移RNA(tRNA)是修饰程度最高的RNA种类。液相色谱与质谱联用(LC-MS / MS)是表征tRNA修饰的强大工具,其中涉及使用碱基特异性核糖核酸对tRNA进行预处理,以产生适合MS的较小寡核苷酸。但是,tRNA的碱基组成会严重影响碱基特异性消化产物的质量和数量。这通常导致序列信息的丢失。在这里,我们报告了一种通过结合LC-MS / MS和RNA-seq技术结合位点特异性RNase H消化以单碱基分辨率对tRNA修饰进行全方位分析的方法。设计关键步骤是为了生成具有最佳长度和电离特性的高质量产品。碰撞能量与碰撞能量之间的线性关系m / z的寡核苷酸显着提高了碰撞诱导解离(CID)光谱的信息含量。使用新的纳入标准收集修改普查结果,误报率高达95%。迄今为止尚未研究的小鼠线粒体tRNA His(GUG)和tRNA Val(UAC)的作图说明了该方法。这些tRNA上的五个修饰物种的身份和位置已得到充分表征。这种方法普遍适用于任何tRNA种类,并提供了从实验上可实现的途径,以具有高序列覆盖率的转录后修饰tRNA从头测序。











































 京公网安备 11010802027423号
京公网安备 11010802027423号