当前位置:
X-MOL 学术
›
Comput. Struct. Biotechnol. J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Improved noninvasive fetal variant calling using standardized benchmarking approaches
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2020-12-31 , DOI: 10.1016/j.csbj.2020.12.032 Tom Rabinowitz , Shira Deri-Rozov , Noam Shomron
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2020-12-31 , DOI: 10.1016/j.csbj.2020.12.032 Tom Rabinowitz , Shira Deri-Rozov , Noam Shomron
|
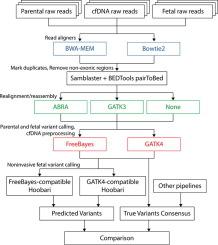
The technology of noninvasive prenatal testing (NIPT) enables risk-free detection of genetic conditions in the fetus, by analysis of cell-free DNA (cfDNA) in maternal blood. For chromosomal abnormalities, NIPT often effectively replaces invasive tests (e.g. amniocentesis), although it is considered as screening rather than diagnostics. Most recently, the NIPT has been applied to genome-wide, comprehensive genotyping of the fetus using cfDNA, i.e. identifying all its genetic variants and mutations. Previously, we suggested that NIPD should be treated as a special case of variant calling, and presented , the first software tool for noninvasive fetal variant calling. Using a unique pipeline, we were able to comprehensively decipher the inheritance of SNPs and indels. A few caveats still exist in this pipeline. Performance was lower for indels and biparental loci (i.e. where both parents carry the same mutation), and performance was not uniform across the genome. Here we utilized standardized methods for benchmarking of variant calling pipelines and applied them to noninvasive fetal variant calling. By using the best performing pipeline and by focusing on coding regions, we showed that noninvasive fetal genotyping greatly improves performance, particularly in indels and biparental loci. These results emphasize the importance of using widely accepted concepts to describe the challenge of genome-wide NIPT of point mutations; and demonstrate a benchmarking process for the first time in this field. This study brings genome-wide and complete NIPD closer to the clinic; while potentially alleviating uncertainty and anxiety during pregnancy, and promoting informed choices among families and physicians.
中文翻译:

使用标准化基准方法改进非侵入性胎儿变异检出
无创产前检测 (NIPT) 技术通过分析母体血液中的游离 DNA (cfDNA),能够无风险地检测胎儿的遗传状况。对于染色体异常,NIPT 通常有效地取代侵入性检测(例如羊膜穿刺术),尽管它被认为是筛查而不是诊断。最近,NIPT 已应用于使用 cfDNA 对胎儿进行全基因组、全面的基因分型,即识别其所有遗传变异和突变。之前,我们建议 NIPD 应被视为变异检出的特殊情况,并提出了第一个用于非侵入性胎儿变异检出的软件工具。使用独特的流程,我们能够全面破译 SNP 和插入缺失的遗传。这条管道中仍然存在一些警告。插入缺失和双亲基因座(即父母双方都携带相同突变)的性能较低,并且整个基因组的性能并不统一。在这里,我们利用标准化方法对变异检出流程进行基准测试,并将其应用于非侵入性胎儿变异检出。通过使用性能最佳的流程并关注编码区域,我们表明非侵入性胎儿基因分型极大地提高了性能,特别是在插入缺失和双亲基因座方面。这些结果强调了使用广泛接受的概念来描述点突变的全基因组 NIPT 挑战的重要性;并首次在该领域展示了基准测试流程。这项研究使全基因组和完整的 NIPD 更接近临床;同时有可能减轻怀孕期间的不确定性和焦虑,并促进家庭和医生做出明智的选择。
更新日期:2020-12-31
中文翻译:

使用标准化基准方法改进非侵入性胎儿变异检出
无创产前检测 (NIPT) 技术通过分析母体血液中的游离 DNA (cfDNA),能够无风险地检测胎儿的遗传状况。对于染色体异常,NIPT 通常有效地取代侵入性检测(例如羊膜穿刺术),尽管它被认为是筛查而不是诊断。最近,NIPT 已应用于使用 cfDNA 对胎儿进行全基因组、全面的基因分型,即识别其所有遗传变异和突变。之前,我们建议 NIPD 应被视为变异检出的特殊情况,并提出了第一个用于非侵入性胎儿变异检出的软件工具。使用独特的流程,我们能够全面破译 SNP 和插入缺失的遗传。这条管道中仍然存在一些警告。插入缺失和双亲基因座(即父母双方都携带相同突变)的性能较低,并且整个基因组的性能并不统一。在这里,我们利用标准化方法对变异检出流程进行基准测试,并将其应用于非侵入性胎儿变异检出。通过使用性能最佳的流程并关注编码区域,我们表明非侵入性胎儿基因分型极大地提高了性能,特别是在插入缺失和双亲基因座方面。这些结果强调了使用广泛接受的概念来描述点突变的全基因组 NIPT 挑战的重要性;并首次在该领域展示了基准测试流程。这项研究使全基因组和完整的 NIPD 更接近临床;同时有可能减轻怀孕期间的不确定性和焦虑,并促进家庭和医生做出明智的选择。











































 京公网安备 11010802027423号
京公网安备 11010802027423号