International Journal for Parasitology: Parasites and Wildlife ( IF 2.0 ) Pub Date : 2020-12-25 , DOI: 10.1016/j.ijppaw.2020.12.005 Remigiusz Gałęcki , Jerzy Jaroszewski , Tadeusz Bakuła , Xuenan Xuan
|
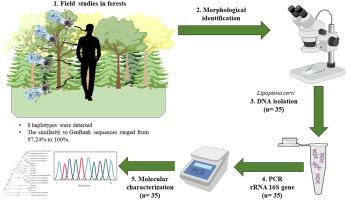
The activity of Lipoptena cervi has intensified in Poland in recent years. The population genetics of this ectoparasite in Poland has never been described in the literature. The objectives of this study were to investigate the population genetics of L. cervi in selected regions of Poland, to evaluate molecular differences between L. cervi populations, and to determine phylogenetic relationships with other L. cervi sequences obtained in previous studies. In 2019, louse flies were sampled in natural mixed forests in five Polish voivodeships. Seven samples of L. cervi were collected from each voivodeship, and a total of 35 insects were analyzed molecularly. In the first step, Lipoptena spp. were identified to species level under a stereoscopic microscope. A fragment of the rRNA 16S gene was used as a marker to identify L. cervi by the PCR assay. The sequences were assigned accession numbers MT337409 to MT337416. A total of eight haplotypes were identified, two of which were dominant. In the obtained sequences, intraspecific pairwise genetic distances varied between 0.000 and 0.0496 (m = 0.0135; SD = 0.0149; SE = 0.0006; V = 110.11). Mean interpopulation diversity was d = 0.0135 (SE = 0.0027). The acquired nucleotide sequences were highly similar to the sequences from the Czech Republic (MF495940, AF322437), Lithuania (MN889542-MN889544) and Poland (MF541726–MF541729). The similarity with GenBank sequences ranged from 97.24% to 100%. This study revealed two dominant haplotypes of L. cervi in Poland, MT337410 and MT337413. Fragments of the analyzed sequences were detected in only one voivodeship. These findings suggest that the two dominant sequences are the oldest sequences that gave rise to the locally identified haplotypes. The lack of significant correlations with the sequences obtained in regions situated west of the research sites suggests the presence of other genetic populations in Europe.
中文翻译:

从波兰收集的环境样品中获得的Liptoptena cervi的分子表征
近年来,波兰的Lipoptena cervi活性增强。在波兰,这种外寄生虫的种群遗传学从未在文献中描述过。这项研究的目的是调查波兰某些地区的宫颈乳杆菌的种群遗传学,评估宫颈乳杆菌种群之间的分子差异,并确定与先前研究中获得的其他宫颈乳杆菌序列的系统发育关系。2019年,在五个波兰省的天然混交林中采集了虱蝇。从每个省收集了7份L. cervi cervi样本,并对35种昆虫进行了分子分析。第一步,Lipoptenaspp。在立体显微镜下被鉴定到物种水平。rRNA 16S基因的片段用作通过PCR测定法鉴定宫颈乳杆菌的标记。序列被分配了登录号MT337409至MT337416。总共鉴定出八种单倍型,其中两种是优势单倍型。在获得的序列中,种内成对遗传距离在0.000和0.0496之间变化(m = 0.0135;SD = 0.0149;SE = 0.0006;V = 110.11)。平均种群多样性为d = 0.0135(SE = 0.0027)。获得的核苷酸序列与捷克共和国(MF495940,AF322437),立陶宛(MN889542-MN889544)和波兰(MF541726–MF541729)的序列高度相似。与GenBank序列的相似性范围为97.24%至100%。这项研究揭示了波兰的L. cervi的两个主要单倍型MT337410和MT337413。仅一次检测到分析序列的片段。这些发现表明,两个显性序列是最古老的序列,产生了局部鉴定的单倍型。与位于研究地点以西的区域中获得的序列缺乏显着相关性,表明在欧洲存在其他遗传种群。











































 京公网安备 11010802027423号
京公网安备 11010802027423号