当前位置:
X-MOL 学术
›
Chem. Res. Toxicol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
DeepDILI: Deep Learning-Powered Drug-Induced Liver Injury Prediction Using Model-Level Representation
Chemical Research in Toxicology ( IF 4.1 ) Pub Date : 2020-12-23 , DOI: 10.1021/acs.chemrestox.0c00374 Ting Li 1, 2 , Weida Tong 1 , Ruth Roberts 1, 3, 4 , Zhichao Liu 1 , Shraddha Thakkar 5
Chemical Research in Toxicology ( IF 4.1 ) Pub Date : 2020-12-23 , DOI: 10.1021/acs.chemrestox.0c00374 Ting Li 1, 2 , Weida Tong 1 , Ruth Roberts 1, 3, 4 , Zhichao Liu 1 , Shraddha Thakkar 5
Affiliation
|
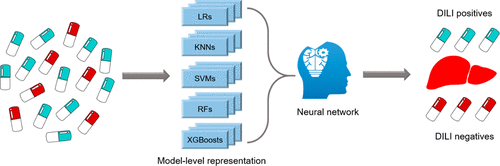
Drug-induced liver injury (DILI) is the most frequently reported single cause of safety-related withdrawal of marketed drugs. It is essential to identify drugs with DILI potential at the early stages of drug development. In this study, we describe a deep learning-powered DILI (DeepDILI) prediction model created by combining model-level representation generated by conventional machine learning (ML) algorithms with a deep learning framework based on Mold2 descriptors. We conducted a comprehensive evaluation of the proposed DeepDILI model performance by posing several critical questions: (1) Could the DILI potential of newly approved drugs be predicted by accumulated knowledge of early approved ones? (2) is model-level representation more informative than molecule-based representation for DILI prediction? and (3) could improved model explainability be established? For question 1, we developed the DeepDILI model using drugs approved before 1997 to predict the DILI potential of those approved thereafter. As a result, the DeepDILI model outperformed the five conventional ML algorithms and two state-of-the-art ensemble methods with a Matthews correlation coefficient (MCC) value of 0.331. For question 2, we demonstrated that the DeepDILI model’s performance was significantly improved (i.e., a MCC improvement of 25.86% in test set) compared with deep neural networks based on molecule-based representation. For question 3, we found 21 chemical descriptors that were enriched, suggesting a strong association with DILI outcome. Furthermore, we found that the DeepDILI model has more discrimination power to identify the DILI potential of drugs belonging to the World Health Organization therapeutic category of ‘alimentary tract and metabolism’. Moreover, the DeepDILI model based on Mold2 descriptors outperformed the ones with Mol2vec and MACCS descriptors. Finally, the DeepDILI model was applied to the recent real-world problem of predicting any DILI concern for potential COVID-19 treatments from repositioning drug candidates. Altogether, this developed DeepDILI model could serve as a promising tool for screening for DILI risk of compounds in the preclinical setting, and the DeepDILI model is publicly available through https://github.com/TingLi2016/DeepDILI.
中文翻译:

DeepDILI:使用模型级表示的深度学习驱动的药物诱发的肝损伤预测
药物诱发的肝损伤(DILI)是最常报告的与安全性相关的撤回市售药物的单一原因。在药物开发的早期阶段,确定具有DILI潜力的药物至关重要。在这项研究中,我们描述了一种深度学习动力的DILI(DeepDILI)预测模型,该模型是通过将常规机器学习(ML)算法生成的模型级表示与基于Mold2描述符的深度学习框架相结合而创建的。我们通过提出几个关键问题对建议的DeepDILI模型性能进行了全面评估:(1)能否通过对早期批准药物的积累知识来预测新批准药物的DILI潜力?(2)在DILI预测中,模型级表示是否比基于分子的表示更具信息性?(3)是否可以建立改进的模型可解释性?对于问题1,我们使用1997年之前批准的药物开发了DeepDILI模型,以预测之后批准的药物的DILI潜力。结果,DeepDILI模型以Matthews相关系数(MCC)值为0.331胜过了五种常规ML算法和两种最新的集成方法。对于问题2,我们证明与基于分子表示的深度神经网络相比,DeepDILI模型的性能得到了显着改善(即,测试集中的MCC改善了25.86%)。对于问题3,我们发现了丰富的21种化学描述符,表明与DILI结局密切相关。此外,我们发现DeepDILI模型具有更多的判别能力,可以识别属于世界卫生组织“消化道和代谢”治疗类别的药物的DILI潜力。此外,基于Mold2描述符的DeepDILI模型优于具有Mol2vec和MACCS描述符的模型。最后,DeepDILI模型应用于最近的现实世界问题,即通过重新定位候选药物来预测DILI对潜在COVID-19治疗的关注。总体而言,这种开发的DeepDILI模型可以作为一种有前途的工具,用于在临床前环境中筛查化合物的DILI风险,并且DeepDILI模型可通过https://github.com/TingLi2016/DeepDILI公开获得。
更新日期:2021-02-15
中文翻译:

DeepDILI:使用模型级表示的深度学习驱动的药物诱发的肝损伤预测
药物诱发的肝损伤(DILI)是最常报告的与安全性相关的撤回市售药物的单一原因。在药物开发的早期阶段,确定具有DILI潜力的药物至关重要。在这项研究中,我们描述了一种深度学习动力的DILI(DeepDILI)预测模型,该模型是通过将常规机器学习(ML)算法生成的模型级表示与基于Mold2描述符的深度学习框架相结合而创建的。我们通过提出几个关键问题对建议的DeepDILI模型性能进行了全面评估:(1)能否通过对早期批准药物的积累知识来预测新批准药物的DILI潜力?(2)在DILI预测中,模型级表示是否比基于分子的表示更具信息性?(3)是否可以建立改进的模型可解释性?对于问题1,我们使用1997年之前批准的药物开发了DeepDILI模型,以预测之后批准的药物的DILI潜力。结果,DeepDILI模型以Matthews相关系数(MCC)值为0.331胜过了五种常规ML算法和两种最新的集成方法。对于问题2,我们证明与基于分子表示的深度神经网络相比,DeepDILI模型的性能得到了显着改善(即,测试集中的MCC改善了25.86%)。对于问题3,我们发现了丰富的21种化学描述符,表明与DILI结局密切相关。此外,我们发现DeepDILI模型具有更多的判别能力,可以识别属于世界卫生组织“消化道和代谢”治疗类别的药物的DILI潜力。此外,基于Mold2描述符的DeepDILI模型优于具有Mol2vec和MACCS描述符的模型。最后,DeepDILI模型应用于最近的现实世界问题,即通过重新定位候选药物来预测DILI对潜在COVID-19治疗的关注。总体而言,这种开发的DeepDILI模型可以作为一种有前途的工具,用于在临床前环境中筛查化合物的DILI风险,并且DeepDILI模型可通过https://github.com/TingLi2016/DeepDILI公开获得。



























 京公网安备 11010802027423号
京公网安备 11010802027423号