当前位置:
X-MOL 学术
›
Neuropathol. Appl. Neurobiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Accurate calling of KIAA1549‐BRAF fusions from DNA of human brain tumours using methylation array‐based copy number and gene panel sequencing data
Neuropathology and Applied Neurobiology ( IF 4.0 ) Pub Date : 2021-01-17 , DOI: 10.1111/nan.12683 Damian Stichel 1, 2 , Daniel Schrimpf 1, 2 , Philipp Sievers 1, 2 , Annekathrin Reinhardt 1, 2 , Abigail K Suwala 1, 2 , Martin Sill 3, 4 , David E Reuss 1, 2 , Andrey Korshunov 1, 2, 3 , Belén M Casalini 1, 2 , Alexander C Sommerkamp 3, 5, 6 , Jonas Ecker 3, 7, 8 , Florian Selt 3, 7, 8 , Dominik Sturm 3, 5, 7 , Astrid Gnekow 9 , Arend Koch 10, 11 , Michèle Simon 12 , Pablo Hernáiz Driever 12 , Ulrich Schüller 13, 14, 15 , David Capper 10, 11 , Cornelis M van Tilburg 3, 7, 8 , Olaf Witt 3, 7, 8 , Till Milde 3, 7, 8 , Stefan M Pfister 3, 4, 7 , David T W Jones 3, 5 , Andreas von Deimling 1, 2 , Felix Sahm 1, 2, 3 , Annika K Wefers 1, 2, 3, 13, 14, 15
Neuropathology and Applied Neurobiology ( IF 4.0 ) Pub Date : 2021-01-17 , DOI: 10.1111/nan.12683 Damian Stichel 1, 2 , Daniel Schrimpf 1, 2 , Philipp Sievers 1, 2 , Annekathrin Reinhardt 1, 2 , Abigail K Suwala 1, 2 , Martin Sill 3, 4 , David E Reuss 1, 2 , Andrey Korshunov 1, 2, 3 , Belén M Casalini 1, 2 , Alexander C Sommerkamp 3, 5, 6 , Jonas Ecker 3, 7, 8 , Florian Selt 3, 7, 8 , Dominik Sturm 3, 5, 7 , Astrid Gnekow 9 , Arend Koch 10, 11 , Michèle Simon 12 , Pablo Hernáiz Driever 12 , Ulrich Schüller 13, 14, 15 , David Capper 10, 11 , Cornelis M van Tilburg 3, 7, 8 , Olaf Witt 3, 7, 8 , Till Milde 3, 7, 8 , Stefan M Pfister 3, 4, 7 , David T W Jones 3, 5 , Andreas von Deimling 1, 2 , Felix Sahm 1, 2, 3 , Annika K Wefers 1, 2, 3, 13, 14, 15
Affiliation
|
AIMS
KIAA1549-BRAF fusions occur in certain brain tumours and provide druggable targets due to a constitutive activation of the MAP-kinase pathway. We introduce workflows for calling the KIAA1549-BRAF fusion from DNA methylation-array-derived copy number as well as DNA panel sequencing data. METHODS
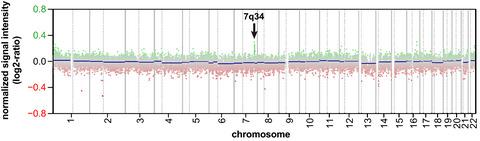
Copy number profiles were analysed by automated screening and visual verification of a tandem duplication on chromosome 7q34, indicative of the KIAA1549-BRAF fusion. Pilocytic astrocytomas of the ICGC cohort with known fusion status were used for validation. KIAA1549-BRAF fusions were called from DNA panel sequencing data using the fusion callers Manta, Arriba with modified filtering criteria and deFuse. We screened DNA methylation and panel sequencing data of 7790 specimens from brain tumour and sarcoma entities. RESULTS
We identified the fusion in 337 brain tumours with both DNA methylation and panel sequencing data. Among these, we detected the fusion from copy number data in 84% and from DNA panel sequencing data in more than 90% using Arriba with modified filters. While in 74% the KIAA1549-BRAF fusion was detected from both methylation-array-derived copy number and panel sequencing data, in 9% it was detected from copy number data only and in 16% from panel data only. The fusion was almost exclusively found in pilocytic astrocytomas, diffuse leptomeningeal glioneuronal tumours and high-grade astrocytomas with piloid features. CONCLUSIONS
The KIAA1549-BRAF fusion can be reliably detected from either DNA methylation array or DNA panel data. The use of both methods is recommended for the most sensitive detection of this diagnostically and therapeutically important marker.
中文翻译:

使用基于甲基化阵列的拷贝数和基因组测序数据从人脑肿瘤的 DNA 中准确调用 KIAA1549-BRAF 融合
AIMS KIAA1549-BRAF 融合发生在某些脑肿瘤中,并且由于 MAP 激酶途径的组成型激活而提供可药物靶点。我们介绍了从 DNA 甲基化阵列衍生的拷贝数以及 DNA 面板测序数据调用 KIAA1549-BRAF 融合的工作流程。方法通过自动筛选和目视验证染色体 7q34 上的串联重复来分析拷贝数谱,表明 KIAA1549-BRAF 融合。具有已知融合状态的 ICGC 队列的毛细胞星形细胞瘤用于验证。KIAA1549-BRAF 融合是使用融合调用程序 Manta、Arriba 和修改后的过滤标准和 deFuse 从 DNA 面板测序数据中调用的。我们筛选了来自脑肿瘤和肉瘤实体的 7790 个样本的 DNA 甲基化和面板测序数据。结果 我们利用 DNA 甲基化和面板测序数据确定了 337 个脑肿瘤的融合。其中,我们使用带有改良过滤器的 Arriba 从 84% 的拷贝数数据和超过 90% 的 DNA 面板测序数据中检测到融合。虽然 74% 的 KIAA1549-BRAF 融合是从甲基化阵列衍生的拷贝数和面板测序数据中检测到的,但 9% 仅从拷贝数数据中检测到,16% 仅从面板数据中检测到。这种融合几乎只见于毛细胞星形细胞瘤、弥漫性软脑膜胶质神经元肿瘤和具有毛状特征的高级别星形细胞瘤。结论 KIAA1549-BRAF 融合可以从 DNA 甲基化阵列或 DNA 面板数据中可靠地检测到。
更新日期:2021-01-17
中文翻译:

使用基于甲基化阵列的拷贝数和基因组测序数据从人脑肿瘤的 DNA 中准确调用 KIAA1549-BRAF 融合
AIMS KIAA1549-BRAF 融合发生在某些脑肿瘤中,并且由于 MAP 激酶途径的组成型激活而提供可药物靶点。我们介绍了从 DNA 甲基化阵列衍生的拷贝数以及 DNA 面板测序数据调用 KIAA1549-BRAF 融合的工作流程。方法通过自动筛选和目视验证染色体 7q34 上的串联重复来分析拷贝数谱,表明 KIAA1549-BRAF 融合。具有已知融合状态的 ICGC 队列的毛细胞星形细胞瘤用于验证。KIAA1549-BRAF 融合是使用融合调用程序 Manta、Arriba 和修改后的过滤标准和 deFuse 从 DNA 面板测序数据中调用的。我们筛选了来自脑肿瘤和肉瘤实体的 7790 个样本的 DNA 甲基化和面板测序数据。结果 我们利用 DNA 甲基化和面板测序数据确定了 337 个脑肿瘤的融合。其中,我们使用带有改良过滤器的 Arriba 从 84% 的拷贝数数据和超过 90% 的 DNA 面板测序数据中检测到融合。虽然 74% 的 KIAA1549-BRAF 融合是从甲基化阵列衍生的拷贝数和面板测序数据中检测到的,但 9% 仅从拷贝数数据中检测到,16% 仅从面板数据中检测到。这种融合几乎只见于毛细胞星形细胞瘤、弥漫性软脑膜胶质神经元肿瘤和具有毛状特征的高级别星形细胞瘤。结论 KIAA1549-BRAF 融合可以从 DNA 甲基化阵列或 DNA 面板数据中可靠地检测到。











































 京公网安备 11010802027423号
京公网安备 11010802027423号