Genomics ( IF 3.4 ) Pub Date : 2020-12-15 , DOI: 10.1016/j.ygeno.2020.12.025 Xuewei Chen 1 , Bencheng Lin 1 , Mingzhu Luo 2 , Wenbin Chu 1 , Ping Li 3 , Huanliang Liu 1 , Zhuge Xi 1 , Rong Fan 4
|
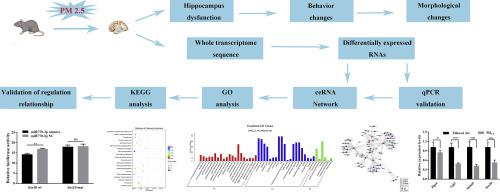
Non-coding RNAs appear to be involved in the regulation of the nervous system. However, no competing endogenous RNA (ceRNA) network related to PM2.5 damage in the hippocampal function has yet been constructed. Herein, we used whole-transcriptome sequencing technology to systematically study the ceRNA network in rat hippocampi after PM2.5 exposure. We identified 100 circRNAs, 67 lncRNAs, 28 miRNAs, and 539 mRNAs and constructed the most comprehensive ceRNA network to date, to our knowledge. Gene Ontology and KEGG analyses showed that the network molecules are involved in synapses, neural projections, and neural development and involve signal pathways such as the synaptic vesicle cycle. Finally, the expression of the differentially expressed RNAs confirmed by quantitative real-time PCR was consistent with the sequencing data. This study systematically dissected the ceRNA atlas related to cognitive memory function in the hippocampal tissue of PM2.5-exposed rats for the first time, to our knowledge, and promotes the development of potential new treatments for cognitive impairment.
中文翻译:

使用 RNA-seq 分析识别暴露于 PM2.5 的大鼠海马中的 circRNA 和 lncRNA 相关 ceRNA 网络
非编码 RNA 似乎参与了神经系统的调节。然而,尚未构建与海马功能中PM 2.5损伤相关的竞争性内源性 RNA (ceRNA) 网络。在此,我们使用全转录组测序技术系统研究了 PM 2.5后大鼠海马中的 ceRNA 网络接触。据我们所知,我们鉴定了 100 个 circRNA、67 个 lncRNA、28 个 miRNA 和 539 个 mRNA,并构建了迄今为止最全面的 ceRNA 网络。基因本体和KEGG分析表明,网络分子参与突触、神经投射和神经发育,并涉及突触小泡循环等信号通路。最后,通过定量实时PCR证实的差异表达RNA的表达与测序数据一致。据我们所知,本研究首次系统剖析了暴露于PM 2.5的大鼠海马组织中与认知记忆功能相关的ceRNA图谱,促进了认知障碍潜在新疗法的开发。









































 京公网安备 11010802027423号
京公网安备 11010802027423号