当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Exploring the Ligand Preferences of the PHD1 Domain of Histone Demethylase KDM5A Reveals Tolerance for Modifications of the Q5 Residue of Histone 3
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2020-12-14 , DOI: 10.1021/acschembio.0c00891 Sarah E Anderson 1 , James E Longbotham 2 , Patrick T O'Kane 1 , Fatima S Ugur 3 , Danica Galonić Fujimori 2, 4, 5 , Milan Mrksich 1, 6, 7
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2020-12-14 , DOI: 10.1021/acschembio.0c00891 Sarah E Anderson 1 , James E Longbotham 2 , Patrick T O'Kane 1 , Fatima S Ugur 3 , Danica Galonić Fujimori 2, 4, 5 , Milan Mrksich 1, 6, 7
Affiliation
|
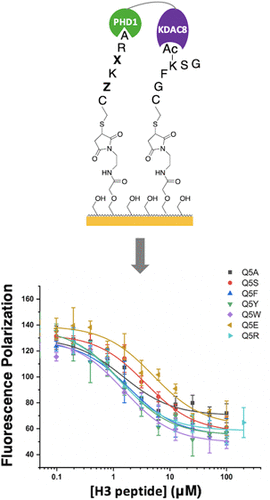
Understanding the ligand preferences of epigenetic reader domains enables identification of modification states of chromatin with which these domains associate and can yield insight into recruitment and catalysis of chromatin-acting complexes. However, thorough exploration of the ligand preferences of reader domains is hindered by the limitations of traditional protein–ligand binding assays. Here, we evaluate the binding preferences of the PHD1 domain of histone demethylase KDM5A using the protein interaction by SAMDI (PI-SAMDI) assay, which measures protein–ligand binding in a high-throughput and sensitive manner via binding-induced enhancement in the activity of a reporter enzyme, in combination with fluorescence polarization. The PI-SAMDI assay was validated by confirming its ability to accurately profile the relative binding affinity of a set of well-characterized histone 3 (H3) ligands of PHD1. The assay was then used to assess the affinity of PHD1 for 361 H3 mutant ligands, a select number of which were further characterized by fluorescence polarization. Together, these experiments revealed PHD1’s tolerance for H3Q5 mutations, including an unexpected tolerance for aromatic residues in this position. Motivated by this finding, we further demonstrate a high-affinity interaction between PHD1 and recently identified Q5-serotonylated H3. This work yields interesting insights into permissible PHD1-H3 interactions and demonstrates the value of interfacing PI-SAMDI and fluorescence polarization in investigations of protein–ligand binding.
中文翻译:

探索组蛋白去甲基化酶 KDM5A 的 PHD1 结构域的配体偏好揭示了对组蛋白 3 Q5 残基修饰的耐受性
了解表观遗传阅读器结构域的配体偏好能够识别与这些结构域相关的染色质的修饰状态,并可以深入了解染色质作用复合物的招募和催化。然而,传统蛋白质-配体结合测定的局限性阻碍了对读者域配体偏好的彻底探索。在这里,我们使用 SAMDI 蛋白质相互作用 (PI-SAMDI) 测定法评估组蛋白去甲基化酶 KDM5A PHD1 结构域的结合偏好,该测定法通过结合诱导的活性增强,以高通量和灵敏的方式测量蛋白质-配体结合。报告酶,与荧光偏振相结合。通过确认 PI-SAMDI 测定能够准确分析一组已充分表征的 PHD1 组蛋白 3 (H3) 配体的相对结合亲和力,对 PI-SAMDI 测定进行了验证。然后使用该测定法评估 PHD1 对 361 个 H3 突变配体的亲和力,其中选定的一些配体通过荧光偏振进一步表征。总之,这些实验揭示了 PHD1 对 H3Q5 突变的耐受性,包括对该位置芳香残基的意外耐受性。受这一发现的启发,我们进一步证明了 PHD1 和最近鉴定的 Q5 血清素化 H3 之间的高亲和力相互作用。这项工作对允许的 PHD1-H3 相互作用产生了有趣的见解,并证明了连接 PI-SAMDI 和荧光偏振在蛋白质-配体结合研究中的价值。
更新日期:2021-01-15
中文翻译:

探索组蛋白去甲基化酶 KDM5A 的 PHD1 结构域的配体偏好揭示了对组蛋白 3 Q5 残基修饰的耐受性
了解表观遗传阅读器结构域的配体偏好能够识别与这些结构域相关的染色质的修饰状态,并可以深入了解染色质作用复合物的招募和催化。然而,传统蛋白质-配体结合测定的局限性阻碍了对读者域配体偏好的彻底探索。在这里,我们使用 SAMDI 蛋白质相互作用 (PI-SAMDI) 测定法评估组蛋白去甲基化酶 KDM5A PHD1 结构域的结合偏好,该测定法通过结合诱导的活性增强,以高通量和灵敏的方式测量蛋白质-配体结合。报告酶,与荧光偏振相结合。通过确认 PI-SAMDI 测定能够准确分析一组已充分表征的 PHD1 组蛋白 3 (H3) 配体的相对结合亲和力,对 PI-SAMDI 测定进行了验证。然后使用该测定法评估 PHD1 对 361 个 H3 突变配体的亲和力,其中选定的一些配体通过荧光偏振进一步表征。总之,这些实验揭示了 PHD1 对 H3Q5 突变的耐受性,包括对该位置芳香残基的意外耐受性。受这一发现的启发,我们进一步证明了 PHD1 和最近鉴定的 Q5 血清素化 H3 之间的高亲和力相互作用。这项工作对允许的 PHD1-H3 相互作用产生了有趣的见解,并证明了连接 PI-SAMDI 和荧光偏振在蛋白质-配体结合研究中的价值。











































 京公网安备 11010802027423号
京公网安备 11010802027423号