当前位置:
X-MOL 学术
›
Bioconjugate Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
One-Pot Synthesis of Defined-Length ssDNA for Multiscaffold DNA Origami
Bioconjugate Chemistry ( IF 4.0 ) Pub Date : 2020-12-13 , DOI: 10.1021/acs.bioconjchem.0c00644 Willem E M Noteborn 1 , Leoni Abendstein 1 , Thomas H Sharp 1
Bioconjugate Chemistry ( IF 4.0 ) Pub Date : 2020-12-13 , DOI: 10.1021/acs.bioconjchem.0c00644 Willem E M Noteborn 1 , Leoni Abendstein 1 , Thomas H Sharp 1
Affiliation
|
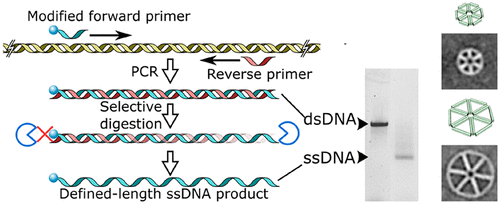
DNA origami nanostructures generally require a single scaffold strand of specific length, combined with many small staple strands. Ideally, the length of the scaffold strand should be dictated by the size of the designed nanostructure. However, synthesizing arbitrary-length single-stranded DNA in sufficient quantities is difficult. Here, we describe a straightforward and accessible method to produce defined-length ssDNA scaffolds using PCR and subsequent selective enzymatic digestion with T7 exonuclease. This approach produced ssDNA with higher yields than other methods and without the need for purification, which significantly decreased the time from PCR to obtaining pure DNA origami. Furthermore, this enabled us to perform true one-pot synthesis of defined-size DNA origami nanostructures. Additionally, we show that multiple smaller ssDNA scaffolds can efficiently substitute longer scaffolds in the formation of DNA origami.
中文翻译:

用于多支架 DNA 折纸的确定长度 ssDNA 的一锅法合成
DNA 折纸纳米结构通常需要特定长度的单个支架链,与许多小主食链相结合。理想情况下,支架链的长度应由设计的纳米结构的尺寸决定。然而,合成足够数量的任意长度的单链 DNA 是很困难的。在这里,我们描述了一种使用 PCR 和随后用 T7 核酸外切酶选择性酶消化来生产定义长度的 ssDNA 支架的简单易行的方法。这种方法产生的 ssDNA 产量比其他方法更高,而且不需要纯化,这显着减少了从 PCR 到获得纯 DNA 折纸的时间。此外,这使我们能够对确定尺寸的 DNA 折纸纳米结构进行真正的一锅法合成。此外,
更新日期:2021-01-20
中文翻译:

用于多支架 DNA 折纸的确定长度 ssDNA 的一锅法合成
DNA 折纸纳米结构通常需要特定长度的单个支架链,与许多小主食链相结合。理想情况下,支架链的长度应由设计的纳米结构的尺寸决定。然而,合成足够数量的任意长度的单链 DNA 是很困难的。在这里,我们描述了一种使用 PCR 和随后用 T7 核酸外切酶选择性酶消化来生产定义长度的 ssDNA 支架的简单易行的方法。这种方法产生的 ssDNA 产量比其他方法更高,而且不需要纯化,这显着减少了从 PCR 到获得纯 DNA 折纸的时间。此外,这使我们能够对确定尺寸的 DNA 折纸纳米结构进行真正的一锅法合成。此外,











































 京公网安备 11010802027423号
京公网安备 11010802027423号