当前位置:
X-MOL 学术
›
Nano Lett.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Quantifying the Dynamics of Protein Self-Organization Using Deep Learning Analysis of Atomic Force Microscopy Data
Nano Letters ( IF 10.8 ) Pub Date : 2020-12-11 , DOI: 10.1021/acs.nanolett.0c03447 Maxim Ziatdinov 1, 2 , Shuai Zhang 3, 4 , Orion Dollar 5 , Jim Pfaendtner 5 , Christopher J. Mundy 4, 5 , Xin Li 1 , Harley Pyles 6, 7 , David Baker 6, 7, 8 , James J. De Yoreo 3, 4 , Sergei V. Kalinin 1
Nano Letters ( IF 10.8 ) Pub Date : 2020-12-11 , DOI: 10.1021/acs.nanolett.0c03447 Maxim Ziatdinov 1, 2 , Shuai Zhang 3, 4 , Orion Dollar 5 , Jim Pfaendtner 5 , Christopher J. Mundy 4, 5 , Xin Li 1 , Harley Pyles 6, 7 , David Baker 6, 7, 8 , James J. De Yoreo 3, 4 , Sergei V. Kalinin 1
Affiliation
|
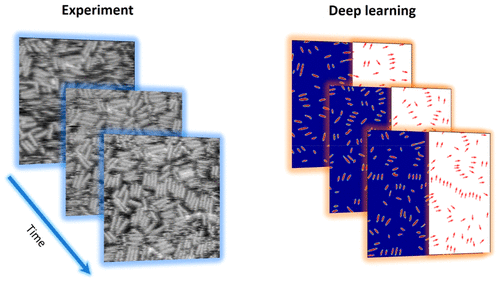
The dynamics of protein self-assembly on the inorganic surface and the resultant geometric patterns are visualized using high-speed atomic force microscopy. The time dynamics of the classical macroscopic descriptors such as 2D fast Fourier transforms, correlation, and pair distribution functions are explored using the unsupervised linear unmixing, demonstrating the presence of static ordered and dynamic disordered phases and establishing their time dynamics. The deep learning (DL)-based workflow is developed to analyze detailed particle dynamics and explore the evolution of local geometries. Finally, we use a combination of DL feature extraction and mixture modeling to define particle neighborhoods free of physics constraints, allowing for a separation of possible classes of particle behavior and identification of the associated transitions. Overall, this work establishes the workflow for the analysis of the self-organization processes in complex systems from observational data and provides insight into the fundamental mechanisms.
中文翻译:

使用原子力显微镜数据的深度学习分析定量蛋白质自组织的动力学
蛋白质在无机表面上自组装的动力学以及由此产生的几何图案可通过高速原子力显微镜观察。使用无监督线性解混方法探索了经典宏观描述符(如2D快速傅里叶变换,相关和对分布函数)的时间动态,证明了静态有序和动态无序相的存在并建立了它们的时间动态。基于深度学习(DL)的工作流旨在分析详细的粒子动力学并探索局部几何形状的演变。最后,我们结合使用DL特征提取和混合建模来定义不受物理约束的粒子邻域,从而允许分离可能的粒子行为类别并识别相关的转变。
更新日期:2021-01-13
中文翻译:

使用原子力显微镜数据的深度学习分析定量蛋白质自组织的动力学
蛋白质在无机表面上自组装的动力学以及由此产生的几何图案可通过高速原子力显微镜观察。使用无监督线性解混方法探索了经典宏观描述符(如2D快速傅里叶变换,相关和对分布函数)的时间动态,证明了静态有序和动态无序相的存在并建立了它们的时间动态。基于深度学习(DL)的工作流旨在分析详细的粒子动力学并探索局部几何形状的演变。最后,我们结合使用DL特征提取和混合建模来定义不受物理约束的粒子邻域,从而允许分离可能的粒子行为类别并识别相关的转变。



























 京公网安备 11010802027423号
京公网安备 11010802027423号