当前位置:
X-MOL 学术
›
ACS Photonics
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Resolving the Sequence of RNA Strands by Tip-Enhanced Raman Spectroscopy
ACS Photonics ( IF 6.5 ) Pub Date : 2020-12-10 , DOI: 10.1021/acsphotonics.0c01486 Zhe He 1 , Weiwei Qiu 2, 3 , Megan E. Kizer 4 , Jizhou Wang 1 , Wencong Chen 5 , Alexei V. Sokolov 1, 3 , Xing Wang 6 , Jonathan Hu 3 , Marlan O. Scully 1, 3
ACS Photonics ( IF 6.5 ) Pub Date : 2020-12-10 , DOI: 10.1021/acsphotonics.0c01486 Zhe He 1 , Weiwei Qiu 2, 3 , Megan E. Kizer 4 , Jizhou Wang 1 , Wencong Chen 5 , Alexei V. Sokolov 1, 3 , Xing Wang 6 , Jonathan Hu 3 , Marlan O. Scully 1, 3
Affiliation
|
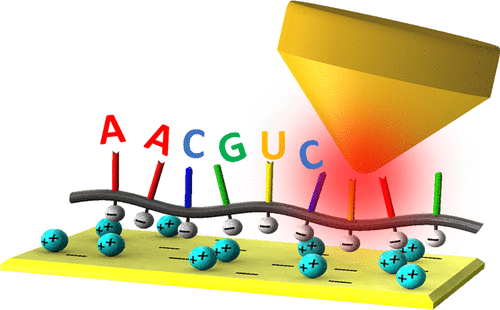
RNA plays critical roles in guiding protein expression and catalyzing biological reactions. The gold standard RNA sequencing method requires converting RNA to complementary DNA (cDNA). This is followed by DNA amplification via polymerase chain reaction (PCR) and sequencing, making RNA sequencing indirect, complicated, and susceptible to sequence data bias. This paper demonstrates RNA imaging at the single-base level while illustrating a direct method to read RNA sequences by tip-enhanced Raman scattering (TERS) technique. To resolve nucleotides within an RNA strand, we adopted gap-mode TERS involving a gold tip and a gold substrate. After analyzing TERS measurements based on the reference sequence, we identified RNA sequences with 90% accuracy. This proof-of-principle RNA imaging method significantly advances a direct RNA sequencing technique without RNA labeling or reverse transcriptase RT-PCR amplification.
中文翻译:

尖端增强拉曼光谱法解析RNA链序列
RNA在指导蛋白质表达和催化生物反应中起关键作用。黄金标准RNA测序方法需要将RNA转换为互补DNA(cDNA)。随后通过聚合酶链反应(PCR)和测序进行DNA扩增,使RNA测序间接,复杂且容易受到序列数据偏差的影响。本文展示了单碱基水平的RNA成像,同时展示了通过尖端增强拉曼散射(TERS)技术读取RNA序列的直接方法。为了解析RNA链中的核苷酸,我们采用了包含金尖端和金底物的缺口模式TERS。在根据参考序列分析TERS测量值后,我们鉴定出具有90%准确性的RNA序列。
更新日期:2021-02-17
中文翻译:

尖端增强拉曼光谱法解析RNA链序列
RNA在指导蛋白质表达和催化生物反应中起关键作用。黄金标准RNA测序方法需要将RNA转换为互补DNA(cDNA)。随后通过聚合酶链反应(PCR)和测序进行DNA扩增,使RNA测序间接,复杂且容易受到序列数据偏差的影响。本文展示了单碱基水平的RNA成像,同时展示了通过尖端增强拉曼散射(TERS)技术读取RNA序列的直接方法。为了解析RNA链中的核苷酸,我们采用了包含金尖端和金底物的缺口模式TERS。在根据参考序列分析TERS测量值后,我们鉴定出具有90%准确性的RNA序列。











































 京公网安备 11010802027423号
京公网安备 11010802027423号