当前位置:
X-MOL 学术
›
Mol. Omics
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
iDHS-DASTS: identifying DNase I hypersensitive sites based on LASSO and stacking learning
Molecular Omics ( IF 3.0 ) Pub Date : 2020-11-12 , DOI: 10.1039/d0mo00115e Shengli Zhang 1 , Zhengpeng Duan 2 , Wenhao Yang 2 , Chenlai Qian 2 , Yiwei You 3
Molecular Omics ( IF 3.0 ) Pub Date : 2020-11-12 , DOI: 10.1039/d0mo00115e Shengli Zhang 1 , Zhengpeng Duan 2 , Wenhao Yang 2 , Chenlai Qian 2 , Yiwei You 3
Affiliation
|
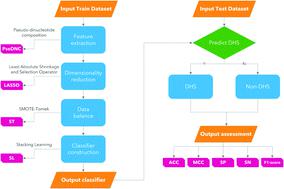
The DNase I hypersensitivity site is an important marker of the DNA regulatory region, and its identification in the DNA sequence is of great significance for biomedical research. However, traditional identification methods are extremely time-consuming and can not obtain an accurate result. In this paper, we proposed a predictor called iDHS-DASTS to identify the DHS based on benchmark datasets. First, we adopt a feature extraction method called PseDNC which can incorporate the original DNA properties and spatial information of the DNA sequence. Then we use a method called LASSO to reduce the dimensions of the original data. Finally, we utilize stacking learning as a classifier, which includes Adaboost, random forest, gradient boosting, extra trees and SVM. Before we train the classifier, we use SMOTE-Tomek to overcome the imbalance of the datasets. In the experiment, our iDHS-DASTS achieves remarkable performance on three benchmark datasets. We achieve state-of-the-art results with over 92.06%, 91.06% and 90.72% accuracy for datasets ![[Doublestruck S]](https://www.rsc.org/images/entities/char_e176.gif) 1,
1, ![[Doublestruck S]](https://www.rsc.org/images/entities/char_e176.gif) 2 and
2 and ![[Doublestruck S]](https://www.rsc.org/images/entities/char_e176.gif) 3, respectively. To verify the validation and transferability of our model, we establish another independent dataset
3, respectively. To verify the validation and transferability of our model, we establish another independent dataset ![[Doublestruck S]](https://www.rsc.org/images/entities/char_e176.gif) 4, for which the accuracy can reach 90.31%. Furthermore, we used the proposed model to construct a user friendly web server called iDHS-DASTS, which is available at http://www.xdu-duan.cn/.
4, for which the accuracy can reach 90.31%. Furthermore, we used the proposed model to construct a user friendly web server called iDHS-DASTS, which is available at http://www.xdu-duan.cn/.
中文翻译:

iDHS-DASTS:基于 LASSO 和堆叠学习识别 DNase I 超敏感位点
DNase I超敏位点是DNA调控区的重要标志物,其在DNA序列中的鉴定对于生物医学研究具有重要意义。然而,传统的识别方法极其耗时且无法获得准确的结果。在本文中,我们提出了一个名为 iDHS-DASTS 的预测器,用于基于基准数据集识别 DHS。首先,我们采用了一种称为 PseDNC 的特征提取方法,它可以结合 DNA 序列的原始 DNA 特性和空间信息。然后我们使用一种叫做 LASSO 的方法对原始数据进行降维。最后,我们利用堆叠学习作为分类器,其中包括 Adaboost、随机森林、梯度提升、额外树和 SVM。在训练分类器之前,我们使用 SMOTE-Tomek 来克服数据集的不平衡。在实验中,我们的 iDHS-DASTS 在三个基准数据集上取得了卓越的性能。我们以超过 92.06%、91.06% 和 90.72% 的数据集准确率获得了最先进的结果![[双重打击 S]](https://www.rsc.org/images/entities/char_e176.gif) 1,
1,![[双重打击 S]](https://www.rsc.org/images/entities/char_e176.gif) 2和
2和![[双重打击 S]](https://www.rsc.org/images/entities/char_e176.gif) 3,分别。为了验证我们模型的有效性和可转移性,我们建立了另一个独立的数据集
3,分别。为了验证我们模型的有效性和可转移性,我们建立了另一个独立的数据集![[双重打击 S]](https://www.rsc.org/images/entities/char_e176.gif) 4,其准确率可以达到 90.31%。此外,我们使用所提出的模型构建了一个名为 iDHS-DASTS 的用户友好的 Web 服务器,该服务器可在 http://www.xdu-duan.cn/ 获得。
4,其准确率可以达到 90.31%。此外,我们使用所提出的模型构建了一个名为 iDHS-DASTS 的用户友好的 Web 服务器,该服务器可在 http://www.xdu-duan.cn/ 获得。
更新日期:2020-12-09
![[Doublestruck S]](https://www.rsc.org/images/entities/char_e176.gif) 1,
1, ![[Doublestruck S]](https://www.rsc.org/images/entities/char_e176.gif) 2 and
2 and ![[Doublestruck S]](https://www.rsc.org/images/entities/char_e176.gif) 3, respectively. To verify the validation and transferability of our model, we establish another independent dataset
3, respectively. To verify the validation and transferability of our model, we establish another independent dataset ![[Doublestruck S]](https://www.rsc.org/images/entities/char_e176.gif) 4, for which the accuracy can reach 90.31%. Furthermore, we used the proposed model to construct a user friendly web server called iDHS-DASTS, which is available at http://www.xdu-duan.cn/.
4, for which the accuracy can reach 90.31%. Furthermore, we used the proposed model to construct a user friendly web server called iDHS-DASTS, which is available at http://www.xdu-duan.cn/.
中文翻译:

iDHS-DASTS:基于 LASSO 和堆叠学习识别 DNase I 超敏感位点
DNase I超敏位点是DNA调控区的重要标志物,其在DNA序列中的鉴定对于生物医学研究具有重要意义。然而,传统的识别方法极其耗时且无法获得准确的结果。在本文中,我们提出了一个名为 iDHS-DASTS 的预测器,用于基于基准数据集识别 DHS。首先,我们采用了一种称为 PseDNC 的特征提取方法,它可以结合 DNA 序列的原始 DNA 特性和空间信息。然后我们使用一种叫做 LASSO 的方法对原始数据进行降维。最后,我们利用堆叠学习作为分类器,其中包括 Adaboost、随机森林、梯度提升、额外树和 SVM。在训练分类器之前,我们使用 SMOTE-Tomek 来克服数据集的不平衡。在实验中,我们的 iDHS-DASTS 在三个基准数据集上取得了卓越的性能。我们以超过 92.06%、91.06% 和 90.72% 的数据集准确率获得了最先进的结果
![[双重打击 S]](https://www.rsc.org/images/entities/char_e176.gif) 1,
1,![[双重打击 S]](https://www.rsc.org/images/entities/char_e176.gif) 2和
2和![[双重打击 S]](https://www.rsc.org/images/entities/char_e176.gif) 3,分别。为了验证我们模型的有效性和可转移性,我们建立了另一个独立的数据集
3,分别。为了验证我们模型的有效性和可转移性,我们建立了另一个独立的数据集![[双重打击 S]](https://www.rsc.org/images/entities/char_e176.gif) 4,其准确率可以达到 90.31%。此外,我们使用所提出的模型构建了一个名为 iDHS-DASTS 的用户友好的 Web 服务器,该服务器可在 http://www.xdu-duan.cn/ 获得。
4,其准确率可以达到 90.31%。此外,我们使用所提出的模型构建了一个名为 iDHS-DASTS 的用户友好的 Web 服务器,该服务器可在 http://www.xdu-duan.cn/ 获得。











































 京公网安备 11010802027423号
京公网安备 11010802027423号