当前位置:
X-MOL 学术
›
Ecol. Evol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A molecular phylogeny of historical and contemporary specimens of an under‐studied micro‐invertebrate group
Ecology and Evolution ( IF 2.3 ) Pub Date : 2020-12-09 , DOI: 10.1002/ece3.7042 Russell J S Orr 1 , Maja M Sannum 1 , Sanne Boessenkool 2 , Emanuela Di Martino 1 , Dennis P Gordon 3 , Hannah L Mello 4 , Matthias Obst 5 , Mali H Ramsfjell 1 , Abigail M Smith 4 , Lee Hsiang Liow 1, 2
Ecology and Evolution ( IF 2.3 ) Pub Date : 2020-12-09 , DOI: 10.1002/ece3.7042 Russell J S Orr 1 , Maja M Sannum 1 , Sanne Boessenkool 2 , Emanuela Di Martino 1 , Dennis P Gordon 3 , Hannah L Mello 4 , Matthias Obst 5 , Mali H Ramsfjell 1 , Abigail M Smith 4 , Lee Hsiang Liow 1, 2
Affiliation
|
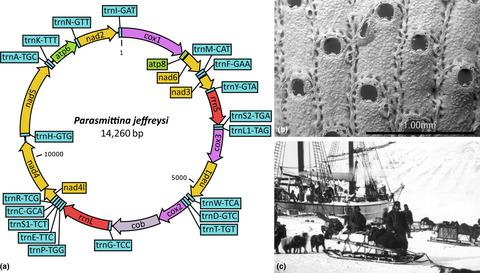
Resolution of relationships at lower taxonomic levels is crucial for answering many evolutionary questions, and as such, sufficiently varied species representation is vital. This latter goal is not always achievable with relatively fresh samples. To alleviate the difficulties in procuring rarer taxa, we have seen increasing utilization of historical specimens in building molecular phylogenies using high throughput sequencing. This effort, however, has mainly focused on large‐bodied or well‐studied groups, with small‐bodied and under‐studied taxa under‐prioritized. Here, we utilize both historical and contemporary specimens, to increase the resolution of phylogenetic relationships among a group of under‐studied and small‐bodied metazoans, namely, cheilostome bryozoans. In this study, we pioneer the sequencing of air‐dried cheilostomes, utilizing a recently developed library preparation method for low DNA input. We evaluate a de novo mitogenome assembly and two iterative methods, using the sequenced target specimen as a reference for mapping, for our sequences. In doing so, we present mitochondrial and ribosomal RNA sequences of 43 cheilostomes representing 37 species, including 14 from historical samples ranging from 50 to 149 years old. The inferred phylogenetic relationships of these samples, analyzed together with publicly available sequence data, are shown in a statistically well‐supported 65 taxa and 17 genes cheilostome tree, which is also the most broadly sampled and largest to date. The robust phylogenetic placement of historical samples whose contemporary conspecifics and/or congenerics have been sequenced verifies the appropriateness of our workflow and gives confidence in the phylogenetic placement of those historical samples for which there are no close relatives sequenced. The success of our workflow is highlighted by the circularization of a total of 27 mitogenomes, seven from historical cheilostome samples. Our study highlights the potential of utilizing DNA from micro‐invertebrate specimens stored in natural history collections for resolving phylogenetic relationships among species.
中文翻译:

正在研究的微型无脊椎动物群的历史和当代标本的分子系统发育
解决较低分类水平上的关系对于回答许多进化问题至关重要,因此,充分多样化的物种代表性至关重要。使用相对新鲜的样品并不总是可以实现后一个目标。为了缓解获取稀有类群的困难,我们发现越来越多地利用历史标本来使用高通量测序构建分子系统发育。然而,这项工作主要集中在大型或经过充分研究的群体,而小型和研究不足的类群则没有得到优先考虑。在这里,我们利用历史和当代标本来提高一组尚未研究的小型后生动物(即唇口动物苔藓动物)之间系统发育关系的分辨率。在这项研究中,我们利用最近开发的低 DNA 输入文库制备方法,率先对风干唇口动物进行测序。我们使用测序的目标样本作为我们的序列作图的参考,评估从头线粒体基因组组装和两种迭代方法。在此过程中,我们展示了代表 37 个物种的 43 种唇口动物的线粒体和核糖体 RNA 序列,其中 14 种来自 50 至 149 岁的历史样本。这些样本的推断系统发育关系,与公开可用的序列数据一起分析,显示在统计上得到充分支持的 65 个分类单元和 17 个基因的唇口虫树中,这也是迄今为止采样最广泛和最大的。 当代同种和/或同属已被测序的历史样本的稳健系统发育布局验证了我们工作流程的适当性,并为那些没有近亲测序的历史样本的系统发育布局提供了信心。我们工作流程的成功体现在总共 27 个线粒体基因组的环化上,其中 7 个来自历史唇口动物样本。我们的研究强调了利用自然历史收藏中存储的微型无脊椎动物标本的 DNA 来解决物种之间的系统发育关系的潜力。
更新日期:2021-01-08
中文翻译:

正在研究的微型无脊椎动物群的历史和当代标本的分子系统发育
解决较低分类水平上的关系对于回答许多进化问题至关重要,因此,充分多样化的物种代表性至关重要。使用相对新鲜的样品并不总是可以实现后一个目标。为了缓解获取稀有类群的困难,我们发现越来越多地利用历史标本来使用高通量测序构建分子系统发育。然而,这项工作主要集中在大型或经过充分研究的群体,而小型和研究不足的类群则没有得到优先考虑。在这里,我们利用历史和当代标本来提高一组尚未研究的小型后生动物(即唇口动物苔藓动物)之间系统发育关系的分辨率。在这项研究中,我们利用最近开发的低 DNA 输入文库制备方法,率先对风干唇口动物进行测序。我们使用测序的目标样本作为我们的序列作图的参考,评估从头线粒体基因组组装和两种迭代方法。在此过程中,我们展示了代表 37 个物种的 43 种唇口动物的线粒体和核糖体 RNA 序列,其中 14 种来自 50 至 149 岁的历史样本。这些样本的推断系统发育关系,与公开可用的序列数据一起分析,显示在统计上得到充分支持的 65 个分类单元和 17 个基因的唇口虫树中,这也是迄今为止采样最广泛和最大的。 当代同种和/或同属已被测序的历史样本的稳健系统发育布局验证了我们工作流程的适当性,并为那些没有近亲测序的历史样本的系统发育布局提供了信心。我们工作流程的成功体现在总共 27 个线粒体基因组的环化上,其中 7 个来自历史唇口动物样本。我们的研究强调了利用自然历史收藏中存储的微型无脊椎动物标本的 DNA 来解决物种之间的系统发育关系的潜力。











































 京公网安备 11010802027423号
京公网安备 11010802027423号