当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Tailoring to Search Engines: Bottom-Up Proteomics with Collision Energies Optimized for Identification Confidence
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2020-12-07 , DOI: 10.1021/acs.jproteome.0c00518 Ágnes Révész 1 , Márton Gyula Milley 1 , Kinga Nagy 1 , Dániel Szabó 1, 2 , Gergő Kalló 3 , Éva Csősz 3 , Károly Vékey 1 , László Drahos 1
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2020-12-07 , DOI: 10.1021/acs.jproteome.0c00518 Ágnes Révész 1 , Márton Gyula Milley 1 , Kinga Nagy 1 , Dániel Szabó 1, 2 , Gergő Kalló 3 , Éva Csősz 3 , Károly Vékey 1 , László Drahos 1
Affiliation
|
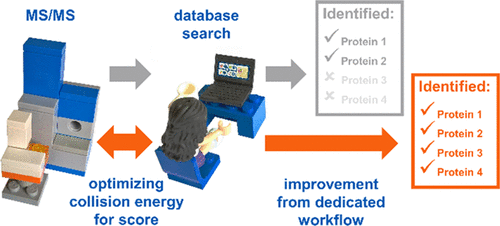
Bottom-up proteomics relies on identification of peptides from tandem mass spectra, usually via matching against sequence databases. Confidence in a peptide–spectrum match can be characterized by a score value given by the database search engines, and it depends on the information content and the quality of the spectrum. The latter are influenced by experimental parameters, of which the collision energy is the most important one in the case of collision-induced dissociation. We examined how the identification score of the Byonic and Andromeda (MaxQuant) engines varies with collision energy for more than a thousand individual peptides from a HeLa tryptic digest on a QTof instrument. We thereby extended our earlier study on Mascot scores and corroborated its findings on the potential bimodal nature of this energy dependence. Optimal energies as a function of m/z show comparable linear trends for the three engines. On the basis of peptide-level results, we designed methods with one or two liquid chromatography–tandem mass spectrometry (LC-MS/MS) runs and various collision energy settings and assessed their practical performance in peptide and protein identification from the HeLa standard sample. A 10–40% gain in various measures, such as the number of identified proteins or sequence coverage, was obtained over the factory default settings. Best performing methods differ for the three engines, suggesting that the experimental parameters should be fine-tuned to the choice of the engine. We also recommend a simple approach and provide reference data to ease the transfer of the optimized methods to other mass spectrometers relevant for proteomics. We demonstrate the utility of this approach on an Orbitrap instrument. Data sets can be accessed via the MassIVE repository (MSV000086379).
中文翻译:

为搜索引擎量身定制:自底向上的蛋白质组学,具有碰撞能量,可优化识别信心
自下而上的蛋白质组学通常依赖于与序列数据库的匹配,从串联质谱中鉴定肽。肽谱匹配的可信度可以通过数据库搜索引擎给出的得分值来表征,它取决于信息内容和质谱图的质量。后者受实验参数的影响,在碰撞诱导的离解中,碰撞能是最重要的。我们检查了QTof仪器上HeLa胰蛋白酶消化物中的1000多种肽段的Byonic and Andromeda(MaxQuant)引擎的识别分数如何随碰撞能量而变化。因此,我们扩展了对Mascot分数的早期研究,并证实了其对这种能量依赖性的潜在双峰性质的发现。m / z显示三个引擎的可比线性趋势。根据多肽水平的结果,我们设计了一种或两种液相色谱-串联质谱(LC-MS / MS)运行方法和各种碰撞能量设置的方法,并评估了它们在HeLa标准样品鉴定多肽和蛋白质中的实际性能。在出厂默认设置下,各种措施(例如,鉴定出的蛋白质数量或序列覆盖率)可提高10-40%。三种发动机的最佳性能方法有所不同,这表明应根据发动机的选择对实验参数进行微调。我们还建议一种简单的方法,并提供参考数据,以简化将优化方法转移到与蛋白质组学相关的其他质谱仪的工作。我们在Orbitrap仪器上演示了此方法的实用性。可以通过MassIVE存储库(MSV000086379)访问数据集。
更新日期:2021-01-01
中文翻译:

为搜索引擎量身定制:自底向上的蛋白质组学,具有碰撞能量,可优化识别信心
自下而上的蛋白质组学通常依赖于与序列数据库的匹配,从串联质谱中鉴定肽。肽谱匹配的可信度可以通过数据库搜索引擎给出的得分值来表征,它取决于信息内容和质谱图的质量。后者受实验参数的影响,在碰撞诱导的离解中,碰撞能是最重要的。我们检查了QTof仪器上HeLa胰蛋白酶消化物中的1000多种肽段的Byonic and Andromeda(MaxQuant)引擎的识别分数如何随碰撞能量而变化。因此,我们扩展了对Mascot分数的早期研究,并证实了其对这种能量依赖性的潜在双峰性质的发现。m / z显示三个引擎的可比线性趋势。根据多肽水平的结果,我们设计了一种或两种液相色谱-串联质谱(LC-MS / MS)运行方法和各种碰撞能量设置的方法,并评估了它们在HeLa标准样品鉴定多肽和蛋白质中的实际性能。在出厂默认设置下,各种措施(例如,鉴定出的蛋白质数量或序列覆盖率)可提高10-40%。三种发动机的最佳性能方法有所不同,这表明应根据发动机的选择对实验参数进行微调。我们还建议一种简单的方法,并提供参考数据,以简化将优化方法转移到与蛋白质组学相关的其他质谱仪的工作。我们在Orbitrap仪器上演示了此方法的实用性。可以通过MassIVE存储库(MSV000086379)访问数据集。











































 京公网安备 11010802027423号
京公网安备 11010802027423号