Journal of Molecular Graphics and Modelling ( IF 2.9 ) Pub Date : 2020-11-27 , DOI: 10.1016/j.jmgm.2020.107816 Shahee Islam 1 , Zarrin Shahzadi 1 , Chaitali Mukhopadhyay 1
|
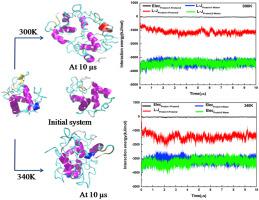
Aggregation of protein causes various diseases including Alzheimer’s disease, Parkinson’s disease, and type II diabetes. It was found that aggregation of protein depends on many factors like temperature, pH, salt type, salt concentration, ionic strength, protein concentration, co solutes. Here we have tried to capture the aggregation mechanism and pathway of hen egg white lysozyme using molecular dynamics simulations at two different temperatures; 300 K and 340 K. Along with the all atom simulations to get the atomistic details of aggregation mechanism, we have used coarse grained simulation with MARTINI force field to monitor the aggregation for longer duration. Our results suggest that due to the aggregation, changes in the conformation of lysozyme are more at 340 K than at 300 K. The change in the conformation of the lysozyme at 300 K is mainly due to aggregation where at 340 K change in conformation of lysozyme is due to both aggregation and temperature. Also, a more compact aggregated system is formed at 340 K.
中文翻译:

溶菌酶的温度依赖性聚集机理和途径:全原子和粗粒分子动力学模拟
蛋白质聚集会导致多种疾病,包括阿尔茨海默氏病,帕金森氏病和II型糖尿病。已经发现蛋白质的聚集取决于许多因素,例如温度,pH,盐类型,盐浓度,离子强度,蛋白质浓度,溶质。在这里,我们试图通过分子动力学模拟在两个不同温度下捕获鸡蛋清溶菌酶的聚集机理和途径。300 K和340K。连同所有原子模拟以获取聚集机制的原子细节,我们使用具有MARTINI力场的粗粒度模拟来监视较长时间的聚集。我们的结果表明,由于聚集,溶菌酶构象的变化在340 K处比在300 K处更大。溶菌酶构象在300K时的变化主要是由于聚集,而在340 K时,溶菌酶的构象在变化是由于聚集和温度。同样,在340 K处形成了更紧凑的聚合系统。



























 京公网安备 11010802027423号
京公网安备 11010802027423号