当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Novel Prokaryotic CRISPR-Cas12a-Based Tool for Programmable Transcriptional Activation and Repression
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2020-11-25 , DOI: 10.1021/acssynbio.0c00424 Christoph Schilling 1 , Mattheos A G Koffas 2, 3 , Volker Sieber 1, 4, 5, 6 , Jochen Schmid 1, 7
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2020-11-25 , DOI: 10.1021/acssynbio.0c00424 Christoph Schilling 1 , Mattheos A G Koffas 2, 3 , Volker Sieber 1, 4, 5, 6 , Jochen Schmid 1, 7
Affiliation
|
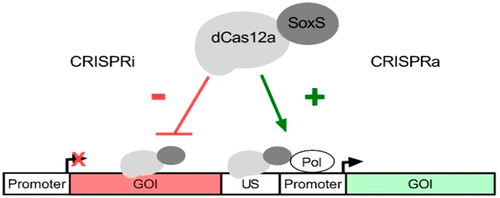
Transcriptional perturbation using inactivated CRISPR-nucleases (dCas) is a common method in eukaryotic organisms. While rare examples of dCas9-based tools for prokaryotes have been described, multiplexing approaches are limited due to the used effector nuclease. For the first time, a dCas12a derived tool for the targeted activation and repression of genes was developed. Therefore, a previously described SoxS activator domain was linked to dCas12a to enable the programmable activation of gene expression. A proof of principle of transcriptional regulation was demonstrated on the basis of fluorescence reporter assays using the alternative host organism Paenibacillus polymyxa as well as Escherichia coli. Single target and multiplex CRISPR interference targeting the exopolysaccharide biosynthesis of P. polymyxa was shown to emulate polymer compositions of gene knockouts. The simultaneous expression of 11 gRNAs targeting multiple lactate dehydrogenases and a butanediol dehydrogenase resulted in decreased lactate formation, as well as an increased butanediol production in microaerobic fermentation processes. Even though Cas12a is more restricted in terms of its genomic target sequences compared to Cas9, its ability to efficiently process its own guide RNAs in vivo makes it a promising tool to orchestrate sophisticated genetic reprogramming of bacterial cells or to screen for engineering targets in the genome. The developed tool will accelerate metabolic engineering efforts in the alternative host organism P. polymyxa and might be also applied for other bacterial cell factories.
中文翻译:

基于 CRISPR-Cas12a 的新型原核工具,用于可编程转录激活和抑制
使用灭活 CRISPR 核酸酶 (dCas) 进行转录扰动是真核生物中的常见方法。虽然已经描述了基于 dCas9 的原核生物工具的罕见例子,但由于使用的效应核酸酶,多重方法受到限制。首次开发了一种用于基因靶向激活和抑制的 dCas12a 衍生工具。因此,之前描述的 SoxS 激活剂结构域与 dCas12a 连接,以实现基因表达的可编程激活。基于使用替代宿主生物多粘类芽孢杆菌和大肠杆菌的荧光报告分析,证明了转录调控原理。针对多粘菌胞外多糖生物合成的单靶点和多重 CRISPR 干扰被证明可以模拟基因敲除的聚合物组合物。靶向多种乳酸脱氢酶和丁二醇脱氢酶的 11 种 gRNA 同时表达,导致微需氧发酵过程中乳酸形成减少,丁二醇产量增加。尽管与 Cas9 相比,Cas12a 在基因组靶序列方面受到更多限制,但其在体内高效处理自身引导 RNA 的能力使其成为协调细菌细胞复杂遗传重编程或筛选基因组中工程靶点的有前途的工具。开发的工具将加速替代宿主生物多粘菌的代谢工程工作,并且也可能应用于其他细菌细胞工厂。
更新日期:2020-12-18
中文翻译:

基于 CRISPR-Cas12a 的新型原核工具,用于可编程转录激活和抑制
使用灭活 CRISPR 核酸酶 (dCas) 进行转录扰动是真核生物中的常见方法。虽然已经描述了基于 dCas9 的原核生物工具的罕见例子,但由于使用的效应核酸酶,多重方法受到限制。首次开发了一种用于基因靶向激活和抑制的 dCas12a 衍生工具。因此,之前描述的 SoxS 激活剂结构域与 dCas12a 连接,以实现基因表达的可编程激活。基于使用替代宿主生物多粘类芽孢杆菌和大肠杆菌的荧光报告分析,证明了转录调控原理。针对多粘菌胞外多糖生物合成的单靶点和多重 CRISPR 干扰被证明可以模拟基因敲除的聚合物组合物。靶向多种乳酸脱氢酶和丁二醇脱氢酶的 11 种 gRNA 同时表达,导致微需氧发酵过程中乳酸形成减少,丁二醇产量增加。尽管与 Cas9 相比,Cas12a 在基因组靶序列方面受到更多限制,但其在体内高效处理自身引导 RNA 的能力使其成为协调细菌细胞复杂遗传重编程或筛选基因组中工程靶点的有前途的工具。开发的工具将加速替代宿主生物多粘菌的代谢工程工作,并且也可能应用于其他细菌细胞工厂。











































 京公网安备 11010802027423号
京公网安备 11010802027423号