Molecular Cell ( IF 14.5 ) Pub Date : 2020-11-25 , DOI: 10.1016/j.molcel.2020.11.014 Meredith Corley 1 , Ryan A Flynn 2 , Byron Lee 2 , Steven M Blue 1 , Howard Y Chang 3 , Gene W Yeo 1
|
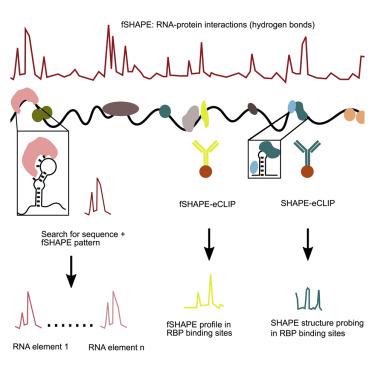
Discovering the interaction mechanism and location of RNA-binding proteins (RBPs) on RNA is critical for understanding gene expression regulation. Here, we apply selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) on in vivo transcripts compared to protein-absent transcripts in four human cell lines to identify transcriptome-wide footprints (fSHAPE) on RNA. Structural analyses indicate that fSHAPE precisely detects nucleobases that hydrogen bond with protein. We demonstrate that fSHAPE patterns predict binding sites of known RBPs, such as iron response elements in both known loci and previously unknown loci in CDC34, SLC2A4RG, COASY, and H19. Furthermore, by integrating SHAPE and fSHAPE with crosslinking and immunoprecipitation (eCLIP) of desired RBPs, we interrogate specific RNA-protein complexes, such as histone stem-loop elements and their nucleotides that hydrogen bond with stem-loop-binding proteins. Together, these technologies greatly expand our ability to study and understand specific cellular RNA interactions in RNA-protein complexes.
中文翻译:

足迹 SHAPE-eCLIP 揭示 RNA-蛋白质界面处转录组范围的氢键
发现 RNA 结合蛋白 (RBP) 在 RNA 上的相互作用机制和位置对于理解基因表达调控至关重要。在这里,我们通过引物延伸 (SHAPE) 对体内转录本进行选择性 2'-羟基酰化分析,与四种人类细胞系中的蛋白质缺失转录本进行比较,以识别 RNA 上的转录组范围足迹 (fSHAPE)。结构分析表明,fSHAPE 能够精确检测与蛋白质形成氢键的核碱基。我们证明 fSHAPE 模式可以预测已知 RBP 的结合位点,例如CDC34 、 SLC2A4RG 、 COASY和H19中已知基因座和先前未知基因座中的铁反应元件。此外,通过将 SHAPE 和 fSHAPE 与所需 RBP 的交联和免疫沉淀 (eCLIP) 相结合,我们研究了特定的 RNA-蛋白质复合物,例如组蛋白茎环元件及其与茎环结合蛋白形成氢键的核苷酸。这些技术共同极大地扩展了我们研究和理解 RNA-蛋白质复合物中特定细胞 RNA 相互作用的能力。











































 京公网安备 11010802027423号
京公网安备 11010802027423号