当前位置:
X-MOL 学术
›
Environ. Sci. Technol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Mercury Methylation Genes Identified across Diverse Anaerobic Microbial Guilds in a Eutrophic Sulfate-Enriched Lake
Environmental Science & Technology ( IF 10.8 ) Pub Date : 2020-11-23 , DOI: 10.1021/acs.est.0c05435 Benjamin D Peterson 1 , Elizabeth A McDaniel 2 , Anna G Schmidt 2 , Ryan F Lepak 1, 3 , Sarah E Janssen 4 , Patricia Q Tran 2, 5 , Robert A Marick 6 , Jacob M Ogorek 4 , John F DeWild 4 , David P Krabbenhoft 4 , Katherine D McMahon 2, 7
Environmental Science & Technology ( IF 10.8 ) Pub Date : 2020-11-23 , DOI: 10.1021/acs.est.0c05435 Benjamin D Peterson 1 , Elizabeth A McDaniel 2 , Anna G Schmidt 2 , Ryan F Lepak 1, 3 , Sarah E Janssen 4 , Patricia Q Tran 2, 5 , Robert A Marick 6 , Jacob M Ogorek 4 , John F DeWild 4 , David P Krabbenhoft 4 , Katherine D McMahon 2, 7
Affiliation
|
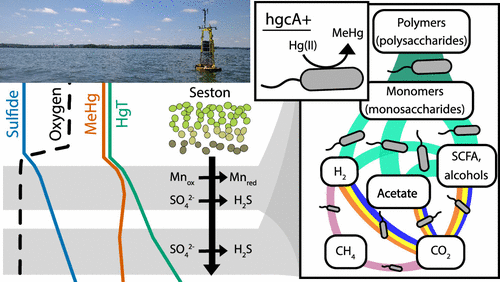
Mercury (Hg) methylation is a microbially mediated process that converts inorganic Hg into bioaccumulative, neurotoxic methylmercury (MeHg). The metabolic activity of methylating organisms is highly dependent on biogeochemical conditions, which subsequently influences MeHg production. However, our understanding of the ecophysiology of methylators in natural ecosystems is still limited. Here, we identified potential locations of MeHg production in the anoxic, sulfidic hypolimnion of a freshwater lake. At these sites, we used shotgun metagenomics to characterize microorganisms with the Hg-methylation gene hgcA. Putative methylators were dominated by hgcA sequences divergent from those in well-studied, confirmed methylators. Using genome-resolved metagenomics, we identified organisms with hgcA (hgcA+) within the Bacteroidetes and the recently described Kiritimatiellaeota phyla. We identified hgcA+ genomes derived from sulfate-reducing bacteria, but these accounted for only 22% of hgcA+ genome coverage. The most abundant hgcA+ genomes were from fermenters, accounting for over half of the hgcA gene coverage. Many of these organisms also mediate hydrolysis of polysaccharides, likely from cyanobacterial blooms. This work highlights the distribution of the Hg-methylation genes across microbial metabolic guilds and indicate that primary degradation of polysaccharides and fermentation may play an important but unrecognized role in MeHg production in the anoxic hypolimnion of freshwater lakes.
中文翻译:

在富含富营养化硫酸盐的湖泊中的不同厌氧微生物群中鉴定出汞甲基化基因
汞 (Hg) 甲基化是一种微生物介导的过程,可将无机汞转化为具有生物蓄积性、具有神经毒性的甲基汞 (MeHg)。甲基化生物的代谢活动高度依赖于生物地球化学条件,进而影响甲基汞的产生。然而,我们对自然生态系统中甲基化因子生态生理学的了解仍然有限。在这里,我们确定了淡水湖缺氧、硫化物下层中可能产生甲基汞的位置。在这些地点,我们使用鸟枪法宏基因组学来表征具有 Hg-甲基化基因hgcA的微生物。假定的甲基化因子以hgcA为主序列与经过充分研究、确认的甲基化器中的序列不同。使用基因组解析的宏基因组学,我们在拟杆菌和最近描述的 Kiritimatiellaeota 门中鉴定了具有hgcA (hgcA+) 的生物体。我们鉴定了源自硫酸盐还原菌的 hgcA+ 基因组,但这些仅占 hgcA+ 基因组覆盖率的 22%。最丰富的 hgcA+ 基因组来自发酵罐,占hgcA的一半以上基因覆盖率。许多这些生物体还介导多糖的水解,可能来自蓝藻水华。这项工作强调了 Hg-甲基化基因在微生物代谢群中的分布,并表明多糖的初级降解和发酵可能在淡水湖泊缺氧低液层中的甲基汞生产中发挥重要但未被认识的作用。
更新日期:2020-12-15
中文翻译:

在富含富营养化硫酸盐的湖泊中的不同厌氧微生物群中鉴定出汞甲基化基因
汞 (Hg) 甲基化是一种微生物介导的过程,可将无机汞转化为具有生物蓄积性、具有神经毒性的甲基汞 (MeHg)。甲基化生物的代谢活动高度依赖于生物地球化学条件,进而影响甲基汞的产生。然而,我们对自然生态系统中甲基化因子生态生理学的了解仍然有限。在这里,我们确定了淡水湖缺氧、硫化物下层中可能产生甲基汞的位置。在这些地点,我们使用鸟枪法宏基因组学来表征具有 Hg-甲基化基因hgcA的微生物。假定的甲基化因子以hgcA为主序列与经过充分研究、确认的甲基化器中的序列不同。使用基因组解析的宏基因组学,我们在拟杆菌和最近描述的 Kiritimatiellaeota 门中鉴定了具有hgcA (hgcA+) 的生物体。我们鉴定了源自硫酸盐还原菌的 hgcA+ 基因组,但这些仅占 hgcA+ 基因组覆盖率的 22%。最丰富的 hgcA+ 基因组来自发酵罐,占hgcA的一半以上基因覆盖率。许多这些生物体还介导多糖的水解,可能来自蓝藻水华。这项工作强调了 Hg-甲基化基因在微生物代谢群中的分布,并表明多糖的初级降解和发酵可能在淡水湖泊缺氧低液层中的甲基汞生产中发挥重要但未被认识的作用。











































 京公网安备 11010802027423号
京公网安备 11010802027423号