当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Single Cell Characterization of a Synthetic Bacterial Clock with a Hybrid Feedback Loop Containing dCas9-sgRNA
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2020-11-24 , DOI: 10.1021/acssynbio.0c00438 John Henningsen 1 , Matthaeus Schwarz-Schilling 1 , Andreas Leibl 1 , Joaquı N Gutiérrez 1 , Sandra Sagredo 1 , Friedrich C Simmel 1
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2020-11-24 , DOI: 10.1021/acssynbio.0c00438 John Henningsen 1 , Matthaeus Schwarz-Schilling 1 , Andreas Leibl 1 , Joaquı N Gutiérrez 1 , Sandra Sagredo 1 , Friedrich C Simmel 1
Affiliation
|
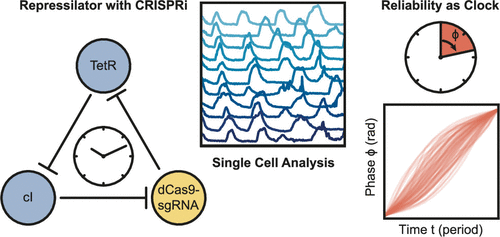
Genetic networks that generate oscillations in gene expression activity are found in a wide range of organisms throughout all kingdoms of life. Oscillatory dynamics facilitates the temporal orchestration of metabolic and growth processes inside cells and organisms, as well as the synchronization of such processes with periodically occurring changes in the environment. Synthetic oscillator gene circuits such as the “repressilator” can perform similar functions in bacteria. Until recently, such circuits were mainly based on a relatively small set of well-characterized transcriptional repressors and activators. A promising, sequence-programmable alternative for gene regulation is given by CRISPR interference (CRISPRi), which enables transcriptional repression of nearly arbitrary gene targets directed by short guide RNA molecules. In order to demonstrate the use of CRISPRi in the context of dynamic gene circuits, we here replaced one of the nodes of a repressilator circuit by the RNA-guided dCas9 protein. Using single cell experiments in microfluidic reactors we show that this system displays robust relaxation oscillations over multiple periods and over several days. With a period of ≈14 bacterial generations, our oscillator is similar in speed as previously reported oscillators. Using an information-theoretic approach for the analysis of the single cell data, the potential of the circuit to act as a synthetic pacemaker for cellular processes is evaluated. We also observe that the oscillator appears to affect cellular growth, leading to variations in growth rate with the oscillator’s frequency.
中文翻译:

具有包含 dCas9-sgRNA 的混合反馈环的合成细菌时钟的单细胞表征
在所有生命领域的多种生物体中都发现了在基因表达活动中产生振荡的遗传网络。振荡动力学促进细胞和生物体内部代谢和生长过程的时间编排,以及这些过程与环境中周期性发生的变化的同步。诸如“抑制器”之类的合成振荡器基因电路可以在细菌中发挥类似的功能。直到最近,此类电路主要基于一组相对较小的、已充分表征的转录抑制子和激活子。 CRISPR 干扰 (CRISPRi) 提供了一种有前途的、序列可编程的基因调控替代方案,它能够在短向导 RNA 分子的指导下对几乎任意的基因靶标进行转录抑制。为了演示 CRISPRi 在动态基因回路中的用途,我们在这里用 RNA 引导的 dCas9 蛋白替换了阻抑器回路的一个节点。在微流体反应器中使用单细胞实验,我们表明该系统在多个时期和几天内表现出强大的弛豫振荡。在约 14 个细菌世代的周期内,我们的振荡器的速度与之前报道的振荡器相似。使用信息论方法分析单细胞数据,评估该电路作为细胞过程的合成起搏器的潜力。我们还观察到振荡器似乎会影响细胞生长,导致生长速率随振荡器频率的变化。
更新日期:2020-12-18
中文翻译:

具有包含 dCas9-sgRNA 的混合反馈环的合成细菌时钟的单细胞表征
在所有生命领域的多种生物体中都发现了在基因表达活动中产生振荡的遗传网络。振荡动力学促进细胞和生物体内部代谢和生长过程的时间编排,以及这些过程与环境中周期性发生的变化的同步。诸如“抑制器”之类的合成振荡器基因电路可以在细菌中发挥类似的功能。直到最近,此类电路主要基于一组相对较小的、已充分表征的转录抑制子和激活子。 CRISPR 干扰 (CRISPRi) 提供了一种有前途的、序列可编程的基因调控替代方案,它能够在短向导 RNA 分子的指导下对几乎任意的基因靶标进行转录抑制。为了演示 CRISPRi 在动态基因回路中的用途,我们在这里用 RNA 引导的 dCas9 蛋白替换了阻抑器回路的一个节点。在微流体反应器中使用单细胞实验,我们表明该系统在多个时期和几天内表现出强大的弛豫振荡。在约 14 个细菌世代的周期内,我们的振荡器的速度与之前报道的振荡器相似。使用信息论方法分析单细胞数据,评估该电路作为细胞过程的合成起搏器的潜力。我们还观察到振荡器似乎会影响细胞生长,导致生长速率随振荡器频率的变化。











































 京公网安备 11010802027423号
京公网安备 11010802027423号