iScience ( IF 4.6 ) Pub Date : 2020-11-24 , DOI: 10.1016/j.isci.2020.101842 Mengying Zhang , Kang Xu , Limei Fu , Qi Wang , Zhenghong Chang , Haozhe Zou , Yan Zhang , Yongsheng Li
|
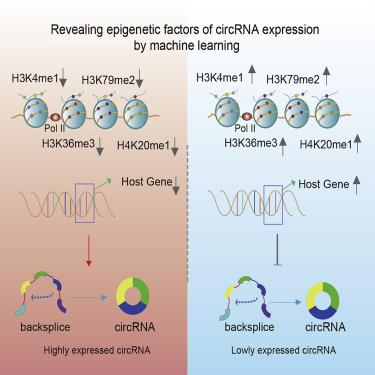
Circular RNAs (circRNAs) have been identified as naturally occurring RNAs that are highly represented in the eukaryotic transcriptome. Although a large number of circRNAs have been reported, the underlying regulatory mechanism of circRNAs biogenesis remains largely unknown. Here, we integrated in-depth multi-omics data including epigenome, transcriptome, and non-coding RNA and identified candidate circRNAs in six cellular contexts. Next, circRNAs were divided into two classes (high versus low) with different expression levels. Machine learning models were constructed that predicted circRNA expression levels based on 11 different histone modifications and host gene expression. We found that the models achieve great accuracy in predicting high versus low expressed circRNAs. Furthermore, the expression levels of host genes of circRNAs, H3k36me3, H3k79me2, and H4k20me1 contributed greatly to the classification models in six cellular contexts. In summary, all these results suggest that epigenetic modifications, particularly histone modifications, can effectively predict expression levels of circRNAs.
中文翻译:

通过机器学习在各种细胞环境中揭示circRNA表达的表观遗传因素
环状RNA(circRNA)已被鉴定为在真核转录组中高度代表的天然存在的RNA。尽管已经报道了许多circRNA,但是circRNA生物发生的潜在调控机制仍然未知。在这里,我们整合了包括表观基因组,转录组和非编码RNA在内的深入多组学数据,并在六个细胞环境中鉴定了候选circRNA。接下来,将circRNA分为具有不同表达水平的两类(高与低)。构建了基于11种不同的组蛋白修饰和宿主基因表达预测circRNA表达水平的机器学习模型。我们发现,该模型在预测高表达和低表达circRNA方面具有很高的准确性。此外,circRNA H3k36me3,H3k79me2和H4k20me1在六个细胞环境中为分类模型做出了巨大贡献。总之,所有这些结果表明表观遗传修饰,特别是组蛋白修饰,可以有效地预测circRNA的表达水平。











































 京公网安备 11010802027423号
京公网安备 11010802027423号