当前位置:
X-MOL 学术
›
Microbiologyopen
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A low‐cost pipeline for soil microbiome profiling
MicrobiologyOpen ( IF 3.9 ) Pub Date : 2020-11-22 , DOI: 10.1002/mbo3.1133 Anita Bollmann-Giolai 1, 2 , Michael Giolai 3 , Darren Heavens 2 , Iain Macaulay 2 , Jacob Malone 1, 4 , Matthew D Clark 3, 4
MicrobiologyOpen ( IF 3.9 ) Pub Date : 2020-11-22 , DOI: 10.1002/mbo3.1133 Anita Bollmann-Giolai 1, 2 , Michael Giolai 3 , Darren Heavens 2 , Iain Macaulay 2 , Jacob Malone 1, 4 , Matthew D Clark 3, 4
Affiliation
|
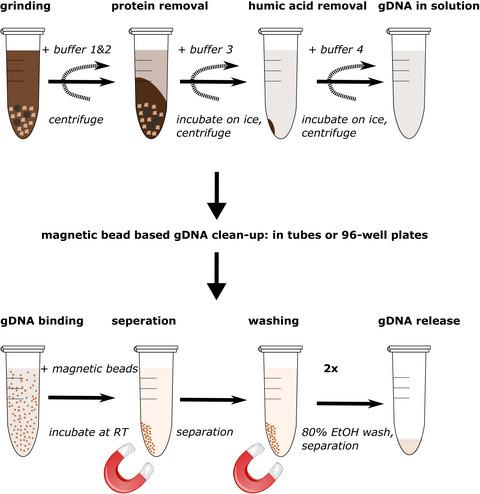
Common bottlenecks in environmental and crop microbiome studies are the consumable and personnel costs necessary for genomic DNA extraction and sequencing library construction. This is harder for challenging environmental samples such as soil, which is rich in Polymerase Chain Reaction (PCR) inhibitors. To address this, we have established a low‐cost genomic DNA extraction method for soil samples. We also present an Illumina‐compatible 16S and ITS rRNA gene amplicon library preparation workflow that uses common laboratory equipment. We evaluated the performance of our genomic DNA extraction method against two leading commercial soil genomic DNA kits (MoBio PowerSoil® and MP Biomedicals™ FastDNA™ SPIN) and a recently published non‐commercial extraction method by Zou et al. (PLoS Biology, 15, e2003916, 2017). Our benchmarking experiment used four different soil types (coniferous, broad‐leafed, and mixed forest plus a standardized cereal crop compost mix) assessing the quality and quantity of the extracted genomic DNA by analyzing sequence variants of 16S V4 and ITS rRNA amplicons. We found that our genomic DNA extraction method compares well to both commercially available genomic DNA extraction kits in DNA quality and quantity. The MoBio PowerSoil® kit, which relies on silica column‐based DNA extraction with extensive washing, delivered the cleanest genomic DNA, for example, best A260:A280 and A260:A230 absorbance ratios. The MP Biomedicals™ FastDNA™ SPIN kit, which uses a large amount of binding material, yielded the most genomic DNA. Our method fits between the two commercial kits, producing both good yields and clean genomic DNA with fragment sizes of approximately 10 kb. Comparative analysis of detected amplicon sequence variants shows that our method correlates well with the two commercial kits. Here, we present a low‐cost genomic DNA extraction method for soil samples that can be coupled to an Illumina‐compatible simple two‐step amplicon library construction workflow for 16S V4 and ITS marker genes. Our method delivers high‐quality genomic DNA at a fraction of the cost of commercial kits and enables cost‐effective, large‐scale amplicon sequencing projects. Notably, our extracted gDNA molecules are long enough to be suitable for downstream techniques such as full gene sequencing or even metagenomics shotgun approaches using long reads (PacBio or Nanopore), 10x Genomics linked reads, and Dovetail genomics.
中文翻译:

用于土壤微生物组分析的低成本管道
环境和作物微生物组研究的常见瓶颈是基因组 DNA 提取和测序文库构建所需的消耗品和人员成本。这对于具有挑战性的环境样品来说更难,例如富含聚合酶链式反应 (PCR) 抑制剂的土壤。为了解决这个问题,我们建立了一种用于土壤样品的低成本基因组 DNA 提取方法。我们还介绍了使用通用实验室设备的 Illumina 兼容的 16S 和 ITS rRNA 基因扩增子文库制备工作流程。我们评估了我们的基因组 DNA 提取方法与两种领先的商业土壤基因组 DNA 试剂盒(MoBio PowerSoil® 和 MP Biomedicals™ FastDNA™ SPIN)以及 Zou 等人最近发表的非商业提取方法的性能。(公共科学图书馆生物学, 15, e2003916, 2017)。我们的基准测试使用四种不同的土壤类型(针叶林、阔叶林和混交林以及标准化的谷类作物堆肥混合物)通过分析 16S V4 和 ITS rRNA 扩增子的序列变体来评估提取的基因组 DNA 的质量和数量。我们发现我们的基因组 DNA 提取方法在 DNA 质量和数量上与市售的两种基因组 DNA 提取试剂盒相媲美。MoBio PowerSoil® 试剂盒依赖于基于硅胶柱的 DNA 提取和大量洗涤,可提供最干净的基因组 DNA,例如最佳的 A260:A280 和 A260:A230 吸光度比。MP Biomedicals™ FastDNA™ SPIN 试剂盒使用大量结合材料,产生最多的基因组 DNA。我们的方法适合两种商业套件,产生良好的产量和干净的基因组 DNA,片段大小约为 10 kb。检测到的扩增子序列变体的比较分析表明,我们的方法与两种商业试剂盒具有很好的相关性。在这里,我们提出了一种用于土壤样品的低成本基因组 DNA 提取方法,该方法可以与 Illumina 兼容的简单两步扩增子文库构建工作流程相结合,用于 16S V4 和 ITS 标记基因。我们的方法以商业试剂盒成本的一小部分提供高质量的基因组 DNA,并实现了具有成本效益的大规模扩增子测序项目。值得注意的是,我们提取的 gDNA 分子足够长,足以适用于下游技术,例如全基因测序,甚至是使用长读长(PacBio 或 Nanopore)、10x Genomics linked read 和 Dovetail 基因组学的宏基因组学鸟枪法。
更新日期:2020-12-23
中文翻译:

用于土壤微生物组分析的低成本管道
环境和作物微生物组研究的常见瓶颈是基因组 DNA 提取和测序文库构建所需的消耗品和人员成本。这对于具有挑战性的环境样品来说更难,例如富含聚合酶链式反应 (PCR) 抑制剂的土壤。为了解决这个问题,我们建立了一种用于土壤样品的低成本基因组 DNA 提取方法。我们还介绍了使用通用实验室设备的 Illumina 兼容的 16S 和 ITS rRNA 基因扩增子文库制备工作流程。我们评估了我们的基因组 DNA 提取方法与两种领先的商业土壤基因组 DNA 试剂盒(MoBio PowerSoil® 和 MP Biomedicals™ FastDNA™ SPIN)以及 Zou 等人最近发表的非商业提取方法的性能。(公共科学图书馆生物学, 15, e2003916, 2017)。我们的基准测试使用四种不同的土壤类型(针叶林、阔叶林和混交林以及标准化的谷类作物堆肥混合物)通过分析 16S V4 和 ITS rRNA 扩增子的序列变体来评估提取的基因组 DNA 的质量和数量。我们发现我们的基因组 DNA 提取方法在 DNA 质量和数量上与市售的两种基因组 DNA 提取试剂盒相媲美。MoBio PowerSoil® 试剂盒依赖于基于硅胶柱的 DNA 提取和大量洗涤,可提供最干净的基因组 DNA,例如最佳的 A260:A280 和 A260:A230 吸光度比。MP Biomedicals™ FastDNA™ SPIN 试剂盒使用大量结合材料,产生最多的基因组 DNA。我们的方法适合两种商业套件,产生良好的产量和干净的基因组 DNA,片段大小约为 10 kb。检测到的扩增子序列变体的比较分析表明,我们的方法与两种商业试剂盒具有很好的相关性。在这里,我们提出了一种用于土壤样品的低成本基因组 DNA 提取方法,该方法可以与 Illumina 兼容的简单两步扩增子文库构建工作流程相结合,用于 16S V4 和 ITS 标记基因。我们的方法以商业试剂盒成本的一小部分提供高质量的基因组 DNA,并实现了具有成本效益的大规模扩增子测序项目。值得注意的是,我们提取的 gDNA 分子足够长,足以适用于下游技术,例如全基因测序,甚至是使用长读长(PacBio 或 Nanopore)、10x Genomics linked read 和 Dovetail 基因组学的宏基因组学鸟枪法。











































 京公网安备 11010802027423号
京公网安备 11010802027423号