iScience ( IF 4.6 ) Pub Date : 2020-11-23 , DOI: 10.1016/j.isci.2020.101841 Shu Yasuda , Nobuyuki Okahashi , Hiroshi Tsugawa , Yusuke Ogata , Kazutaka Ikeda , Wataru Suda , Hiroyuki Arai , Masahira Hattori , Makoto Arita
|
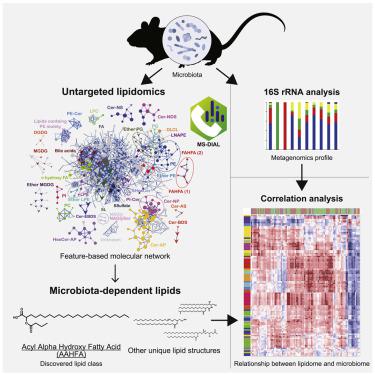
Host-microbiota interactions create a unique metabolic milieu that modulates intestinal environments. Integration of 16S ribosomal RNA (rRNA) sequences and mass spectrometry (MS)-based lipidomics has a great potential to reveal the relationship between bacterial composition and the complex metabolic network in the gut. In this study, we conducted untargeted lipidomics followed by a feature-based molecular MS/MS spectral networking to characterize gut bacteria-dependent lipid subclasses in mice. An estimated 24.8% of lipid molecules in feces were microbiota-dependent, as judged by > 10-fold decrease in antibiotic-treated mice. Among these, there was a series of unique and microbiota-related lipid structures, including acyl alpha-hydroxyl fatty acid (AAHFA) that was newly identified in this study. Based on the integrated analysis of 985 lipid profiles and 16S rRNA sequence data providing 2,494 operational taxonomic units, we could successfully predict the bacterial species responsible for the biosynthesis of these unique lipids, including AAHFA.
中文翻译:

使用LC-MS / MS和16S rRNA序列分析阐明肠道菌群相关脂质
宿主菌群相互作用产生了独特的代谢环境,可调节肠道环境。16S核糖体RNA(rRNA)序列和基于质谱(MS)的脂质组学的整合具有揭示细菌组成与肠道复杂代谢网络之间关系的巨大潜力。在这项研究中,我们进行了无针对性的脂质组学,随后进行了基于特征的分子MS / MS光谱网络分析,以表征小鼠中与肠道细菌有关的脂质亚类。粪便中估计有24.8%的脂质分子是微生物群依赖性的,这可以通过用抗生素治疗的小鼠减少10倍以上来判断。其中,有一系列独特且与微生物群有关的脂质结构,包括在这项研究中新发现的酰基α-羟基脂肪酸(AAHFA)。 基于对985个脂质谱和16S rRNA序列数据的综合分析,提供了2,494个操作生物分类单位,我们可以成功预测出负责这些独特脂质(包括AAHFA)生物合成的细菌物种。











































 京公网安备 11010802027423号
京公网安备 11010802027423号