当前位置:
X-MOL 学术
›
J. Raman Spectrosc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Surface enhanced Raman spectroscopy‐based evaluation of the membrane protein composition of the organohalide‐respiring Sulfurospirillum multivorans
Journal of Raman Spectroscopy ( IF 2.4 ) Pub Date : 2020-11-18 , DOI: 10.1002/jrs.6029 Dana Cialla‐May 1, 2 , Jennifer Gadkari 3 , Andreea Winterfeld 1, 2 , Uwe Hübner 1 , Karina Weber 1, 2 , Gabriele Diekert 3 , Torsten Schubert 3 , Tobias Goris 3, 4 , Jürgen Popp 1, 2
Journal of Raman Spectroscopy ( IF 2.4 ) Pub Date : 2020-11-18 , DOI: 10.1002/jrs.6029 Dana Cialla‐May 1, 2 , Jennifer Gadkari 3 , Andreea Winterfeld 1, 2 , Uwe Hübner 1 , Karina Weber 1, 2 , Gabriele Diekert 3 , Torsten Schubert 3 , Tobias Goris 3, 4 , Jürgen Popp 1, 2
Affiliation
|
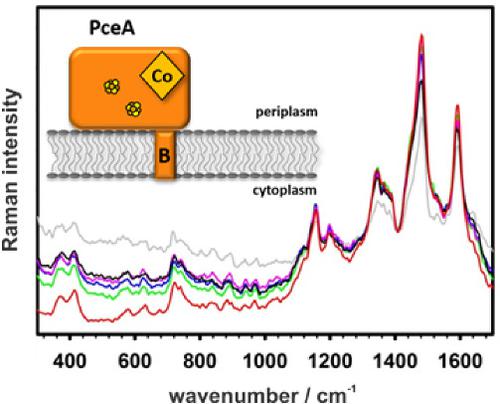
Bacteria often employ different respiratory chains that comprise membrane proteins equipped with various cofactors. Monitoring the protein inventory that is present in the cells under a given cultivation condition is often difficult and time‐consuming. One example of a metabolically versatile bacterium is the microaerophilic organohalide‐respiring Sulfurospirillum multivorans. Here, we used surface enhanced Raman spectroscopy (SERS) to quickly identify the cofactors involved in the respiration of S. multivorans. We cultured the organism with either tetrachloroethene (perchloroethylene, PCE), fumarate, nitrate, or oxygen as electron acceptors. Because the corresponding terminal reductases of the four different respiratory chains harbor different cofactors, specific fingerprint signals in SERS were expected. Silver nanostructures fabricated by means of electron beam lithography were coated with the membrane fractions extracted from the four S. multivorans cultivations, and SERS spectra were recorded. In the case of S. multivorans cultivated with PCE, the recorded SERS spectra were dominated by Raman peaks specific for Vitamin B12. This is attributed to the high abundance of the PCE reductive dehalogenase (PceA), the key enzyme in PCE respiration. After cultivation with oxygen, fumarate, or nitrate, no Raman spectral features of B12 were found.
中文翻译:

基于表面增强拉曼光谱的呼吸器官中有机卤化物磺胺螺菌膜蛋白组成的评估
细菌通常采用不同的呼吸链,这些呼吸链包含配备有各种辅因子的膜蛋白。在给定的培养条件下,监测细胞中存在的蛋白质库存通常是困难且费时的。一种可代谢的通用细菌的例子是可呼吸微卤化的有机卤化物的多伏硫螺旋藻。在这里,我们使用了表面增强拉曼光谱(SERS)来快速识别参与多食链霉菌呼吸作用的辅因子。。我们用四氯乙烯(全氯乙烯,PCE),富马酸酯,硝酸盐或氧气作为电子受体来培养生物。由于四个不同呼吸链的相应末端还原酶带有不同的辅因子,因此可以预期SERS中存在特定的指纹信号。通过电子束光刻技术制备的银纳米结构被覆有从4种多壁链球菌培养物中提取的膜级分,并记录了SERS光谱。对于用PCE培养的多菌链霉菌,记录的SERS光谱以维生素B 12特有的拉曼峰为主。这归因于PCE呼吸中的关键酶PCE还原脱卤素酶(PceA)的丰度很高。用氧气,富马酸盐或硝酸盐培养后,未发现B 12的拉曼光谱特征。
更新日期:2020-11-18
中文翻译:

基于表面增强拉曼光谱的呼吸器官中有机卤化物磺胺螺菌膜蛋白组成的评估
细菌通常采用不同的呼吸链,这些呼吸链包含配备有各种辅因子的膜蛋白。在给定的培养条件下,监测细胞中存在的蛋白质库存通常是困难且费时的。一种可代谢的通用细菌的例子是可呼吸微卤化的有机卤化物的多伏硫螺旋藻。在这里,我们使用了表面增强拉曼光谱(SERS)来快速识别参与多食链霉菌呼吸作用的辅因子。。我们用四氯乙烯(全氯乙烯,PCE),富马酸酯,硝酸盐或氧气作为电子受体来培养生物。由于四个不同呼吸链的相应末端还原酶带有不同的辅因子,因此可以预期SERS中存在特定的指纹信号。通过电子束光刻技术制备的银纳米结构被覆有从4种多壁链球菌培养物中提取的膜级分,并记录了SERS光谱。对于用PCE培养的多菌链霉菌,记录的SERS光谱以维生素B 12特有的拉曼峰为主。这归因于PCE呼吸中的关键酶PCE还原脱卤素酶(PceA)的丰度很高。用氧气,富马酸盐或硝酸盐培养后,未发现B 12的拉曼光谱特征。









































 京公网安备 11010802027423号
京公网安备 11010802027423号