Journal of Computational Physics ( IF 3.8 ) Pub Date : 2020-11-13 , DOI: 10.1016/j.jcp.2020.109999 Agnieszka Borowska , Diana Giurghita , Dirk Husmeier
|
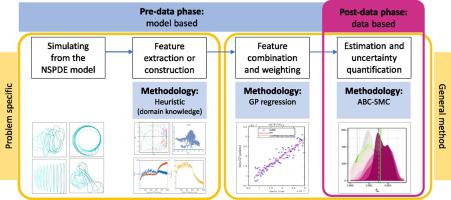
Chemotaxis is a type of cell movement in response to a chemical stimulus which plays a key role in multiple biophysical processes, such as embryogenesis and wound healing, and which is crucial for understanding metastasis in cancer research. In the literature, chemotaxis has been modelled using biophysical models based on systems of nonlinear stochastic partial differential equations (NSPDEs), which are known to be challenging for statistical inference due to the intractability of the associated likelihood and the high computational costs of their numerical integration. Therefore, data analysis in this context has been limited to comparing predictions from NSPDE models to laboratory data using simple descriptive statistics. We present a statistically rigorous framework for parameter estimation in complex biophysical systems described by NSPDEs such as the one of chemotaxis. We adopt a likelihood-free approach based on approximate Bayesian computations with sequential Monte Carlo (ABC-SMC) which allows for circumventing the intractability of the likelihood. To find informative summary statistics, crucial for the performance of ABC, we propose to use a Gaussian process (GP) regression model. The interpolation provided by the GP regression turns out useful on its own merits: it relatively accurately estimates the parameters of the NSPDE model and allows for uncertainty quantification, at a very low computational cost. Our proposed methodology allows for a considerable part of computations to be completed before having observed any data, providing a practical toolbox to experimental scientists whose modes of operation frequently involve experiments and inference taking place at distinct points in time. In an application to externally provided synthetic data we demonstrate that the correction provided by ABC-SMC is essential for accurate estimation of some of the NSPDE model parameters and for more flexible uncertainty quantification.
中文翻译:

高斯过程增强的半自动近似贝叶斯计算:趋化性随机微分方程系统中的参数推断
趋化性是响应化学刺激而发生的一种细胞运动,化学刺激在多种生物物理过程(例如胚胎发生和伤口愈合)中起关键作用,对于理解癌症研究中的转移至关重要。在文献中,已经使用基于非线性随机偏微分方程(NSPDE)系统的生物物理模型对趋化性进行建模,由于相关似然性的难处理性及其数值积分的高计算成本,已知该方法对于统计推断具有挑战性。因此,在这种情况下,数据分析仅限于使用简单的描述性统计将NSPDE模型的预测与实验室数据进行比较。我们提出了统计上严格的框架,用于由NSPDE(例如趋化性之一)描述的复杂生物物理系统中的参数估计。我们采用基于近似贝叶斯计算和顺序蒙特卡洛(ABC-SMC)的无可能性方法,该方法可以避免可能性的难处理性。为了找到对ABC的性能至关重要的信息摘要统计数据,我们建议使用高斯过程(GP)回归模型。GP回归提供的插值本身就很有用:它可以相对准确地估算NSPDE模型的参数,并以非常低的计算成本进行不确定性量化。我们提出的方法可以在观察任何数据之前完成相当一部分计算,向实验科学家提供实用的工具箱,这些科学家的操作模式经常涉及在不同时间点进行的实验和推理。在对外部提供的合成数据的应用中,我们证明了ABC-SMC提供的校正对于准确估算某些NSPDE模型参数和更灵活地进行不确定性量化至关重要。











































 京公网安备 11010802027423号
京公网安备 11010802027423号