Journal of Asia-Pacific Entomology ( IF 1.1 ) Pub Date : 2020-11-11 , DOI: 10.1016/j.aspen.2020.11.003 Mahin Khoshraftar , Javad Nazemi-Rafie , Hamed Ghobari
|
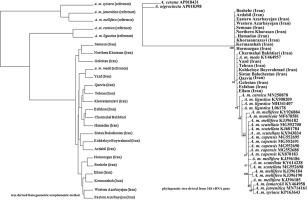
Mitochondrial DNA sequence variations and the geometric morphometric method can be used to differentiate honeybee subspecies and evolutionary lineages. Molecular markers are powerful tools for discriminating honeybee subspecies. In this study, 19 beekeeping sites were selected to collect Iranian honeybee samples. The honeybee forewing images stored at Oberursel (the Bee Data Bank) were used to compare with those of Iranian honeybees using the geometric morphometric method. Furthermore, the abilities of DNA markers to differentiate Iranian honeybees (A. m. meda) from the most common commercial subspecies (A. m. carnica and A. m. ligustica) were assessed. In the present research, 16S rDNA (Mitochondrial 16S rDNA Region) showed greater ability in differentiating Iranian honeybees from other subspecies compared with ATP 6 and 8 and Cyt b. The phylogenetic tree derived from 16S rDNA differentiated A. m. carnica and A. m. ligustica from Iranian honeybees. Principle component analysis (PCA) discriminated C lineage and Z subgroup from A and M lineages using 16S rDNA. In addition, the phylogenetic tree of the 16S rDNA affirmed the findings of the cluster analysis derived from the geometric morphometric method in differentiating A. m. carnica and A. m. ligustica from Iranian honeybees. The cluster analysis grouped reference subspecies of A. m. meda with Iranian honeybees. Moreover, the Discriminant Function Analyses (DFA) differentiated Iranian honeybees from A. m. ligustica and A. m. carnica.
中文翻译:

线粒体ATP 6和8,细胞色素b和16S的疗效评价rDNA基因伊朗蜜蜂的分化(意大利蜜蜂MEDA)从商业亚种蜂L.并用几何形态的方法的比较
线粒体DNA序列变异和几何形态学方法可用于区分蜜蜂亚种和进化谱系。分子标记是区分蜜蜂亚种的有力工具。在这项研究中,选择了19个养蜂场收集伊朗蜜蜂样品。使用几何形态计量学方法,将存储在Oberursel(蜜蜂数据库)的蜜蜂预告图像与伊朗蜜蜂的图像进行比较。此外,DNA标记物能够区分伊朗蜜蜂(A. m。meda)和最常见的商业亚种(A. m。carnica和A. m。ligustica))进行了评估。在本研究中,与ATP 6、8和Cyt b相比,16S rDNA(线粒体16S rDNA区域)显示出更大的区分伊朗蜜蜂与其他亚种的能力。从16S rDNA衍生的系统进化树区分了A. m。卡尼卡和米 ligustica来自伊朗的蜜蜂。主成分分析(PCA)使用16S rDNA从A和M谱系中区分出C谱系和Z子组。此外,16S rDNA的系统发育树证实了从几何形态学方法衍生出的聚类分析结果的判别结果。卡尼卡和米 ligustica来自伊朗的蜜蜂。聚类分析将A.m.的参考亚种分组。梅达与伊朗蜜蜂。此外,判别函数分析(DFA)区分了伊朗蜜蜂和A. m.。女贞和A. m。卡尼卡(Carnica)。











































 京公网安备 11010802027423号
京公网安备 11010802027423号