Journal of Neuroscience Methods ( IF 2.7 ) Pub Date : 2020-11-04 , DOI: 10.1016/j.jneumeth.2020.108988 Abubakhari Sserwadda 1 , Islem Rekik 2
|
Background
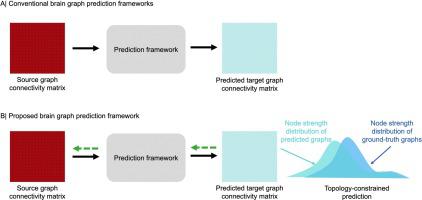
There is a growing need for analyzing medical data such as brain connectomes. However, the unavailability of large-scale training samples increases risks of model over-fitting. Recently, deep learning (DL) architectures quickly gained momentum in synthesizing medical data. However, such frameworks are primarily designed for Euclidean data (e.g., images), overlooking geometric data (e.g., brain connectomes). A few existing geometric DL works that aimed to predict a target brain connectome from a source one primarily focused on domain alignment and were agnostic to preserving the connectome topology.
New method
To address the above limitations, firstly, we adapt the graph translation generative adversarial network (GT GAN) architecture to brain connectomic data. Secondly, we extend the baseline GT GAN to a cyclic graph translation (CGT) GAN, allowing bidirectional brain network translation between the source and target views. Finally, to preserve the topological strength of brain regions of interest (ROIs), we impose a topological strength constraint on the CGT GAN learning, thereby introducing CGTS GAN architecture.
Comparison with existing methods
We compared CGTS with graph translation methods and its ablated versions.
Results
Our deep graph network outperformed the baseline comparison method and its ablated versions in mean squared error (MSE) using multiview autism spectrum disorder connectomic dataset.
Conclusion
We designed a topology-aware bidirectional brain connectome synthesis framework rooted in geometric deep learning, which can be used for data augmentation in clinical diagnosis.
中文翻译:

使用几何深度学习的拓扑指导的循环脑连通性生成
背景
越来越需要分析医学数据,例如脑连接套。但是,无法获得大规模训练样本会增加模型过度拟合的风险。最近,深度学习(DL)架构在合成医学数据方面迅速获得发展势头。但是,这样的框架主要是为欧几里得数据(例如图像),俯视几何数据(例如大脑连接组)而设计的。现有的一些几何DL作品旨在从源头预测目标大脑连接组,该源主要关注域对齐,并且与保留连接组拓扑结构无关。
新方法
为了解决上述局限性,首先,我们将图转换生成对抗网络(GT GAN)架构调整为适用于大脑连接组数据。其次,我们将基线GT GAN扩展为循环图转换(CGT)GAN,从而允许在源视图和目标视图之间进行双向脑网络转换。最后,为了保留感兴趣的大脑区域(ROI)的拓扑强度,我们对CGT GAN学习施加了拓扑强度约束,从而介绍了CGTS GAN体系结构。
与现有方法的比较
我们将CGTS与图翻译方法及其删节版本进行了比较。
结果
我们的深图网络使用多视图自闭症谱系障碍连接数据集的均方误差(MSE)优于基线比较方法及其简化版本。
结论
我们设计了一种基于几何深度学习的拓扑感知双向脑部连接基因组合成框架,该框架可用于临床诊断中的数据增强。











































 京公网安备 11010802027423号
京公网安备 11010802027423号