Journal of Molecular Biology ( IF 4.7 ) Pub Date : 2020-11-03 , DOI: 10.1016/j.jmb.2020.10.030 Nabeel Ahmed 1 , Ulrike A Friedrich 2 , Pietro Sormanni 3 , Prajwal Ciryam 3 , Naomi S Altman 4 , Bernd Bukau 2 , Günter Kramer 2 , Edward P O'Brien 5
|
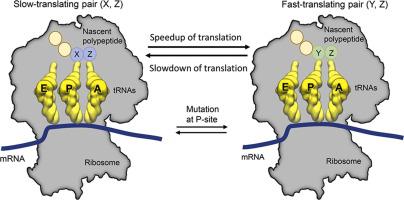
Variation in translation-elongation kinetics along a transcript’s coding sequence plays an important role in the maintenance of cellular protein homeostasis by regulating co-translational protein folding, localization, and maturation. Translation-elongation speed is influenced by molecular factors within mRNA and protein sequences. For example, the presence of proline in the ribosome’s P- or A-site slows down translation, but the effect of other pairs of amino acids, in the context of all 400 possible pairs, has not been characterized. Here, we study Saccharomyces cerevisiae using a combination of bioinformatics, mutational experiments, and evolutionary analyses, and show that many different pairs of amino acids and their associated tRNA molecules predictably and causally encode translation rate information when these pairs are present in the A- and P-sites of the ribosome independent of other factors known to influence translation speed including mRNA structure, wobble base pairing, tripeptide motifs, positively charged upstream nascent chain residues, and cognate tRNA concentration. The fast-translating pairs of amino acids that we identify are enriched four-fold relative to the slow-translating pairs across Saccharomyces cerevisiae’s proteome, while the slow-translating pairs are enriched downstream of domain boundaries. Thus, the chemical identity of amino acid pairs contributes to variability in translation rates, elongation kinetics are causally encoded in the primary structure of proteins, and signatures of evolutionary selection indicate their potential role in co-translational processes.
中文翻译:

核糖体 P 位点和 A 位点的氨基酸对可预测地调节翻译延伸率
沿着转录本编码序列的翻译-延伸动力学变化通过调节共翻译蛋白质折叠、定位和成熟,在维持细胞蛋白质稳态中起着重要作用。翻译延伸速度受 mRNA 和蛋白质序列中的分子因素影响。例如,核糖体 P 位点或 A 位点中脯氨酸的存在会减慢翻译速度,但在所有 400 对可能的氨基酸对的背景下,其他氨基酸对的影响尚未得到表征。在这里,我们研究酿酒酵母使用生物信息学、突变实验和进化分析的组合,并表明当这些对存在于核糖体的 A 位点和 P 位点时,许多不同的氨基酸对及其相关的 tRNA 分子可预测和因果地编码翻译率信息独立于其他已知影响翻译速度的因素,包括 mRNA 结构、摆动碱基配对、三肽基序、带正电荷的上游新生链残基和同源 tRNA 浓度。我们鉴定的快速翻译氨基酸对相对于酿酒酵母中的慢翻译氨基酸对富集了四倍的蛋白质组,而缓慢翻译的对在域边界的下游富集。因此,氨基酸对的化学特性导致翻译速率的变异性,延伸动力学在蛋白质的一级结构中被因果编码,进化选择的特征表明它们在共翻译过程中的潜在作用。











































 京公网安备 11010802027423号
京公网安备 11010802027423号