当前位置:
X-MOL 学术
›
Hum. Mutat.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Efficient detection of copy‐number variations using exome data: Batch‐ and sex‐based analyses
Human Mutation ( IF 3.3 ) Pub Date : 2020-11-01 , DOI: 10.1002/humu.24129 Yuri Uchiyama 1, 2 , Daisuke Yamaguchi 3 , Kazuhiro Iwama 2, 4 , Satoko Miyatake 2, 5 , Kohei Hamanaka 2 , Naomi Tsuchida 1, 2 , Hiromi Aoi 2, 6 , Yoshiteru Azuma 2 , Toshiyuki Itai 2 , Ken Saida 2 , Hiromi Fukuda 2, 7 , Futoshi Sekiguchi 2 , Tomohiro Sakaguchi 2 , Ming Lei 2 , Sachiko Ohori 2 , Masamune Sakamoto 2, 4 , Mitsuhiro Kato 8 , Takayoshi Koike 9 , Yukitoshi Takahashi 9 , Koichi Tanda 10 , Yuki Hyodo 11 , Rachel S Honjo 12 , Debora Romeo Bertola 12 , Chong Ae Kim 12 , Masahide Goto 13 , Tetsuya Okazaki 14 , Hiroyuki Yamada 14 , Yoshihiro Maegaki 14 , Hitoshi Osaka 13 , Lock-Hock Ngu 15 , Ch'ng G Siew 15 , Keng W Teik 15 , Manami Akasaka 16 , Hiroshi Doi 7 , Fumiaki Tanaka 7 , Tomohide Goto 17 , Long Guo 18 , Shiro Ikegawa 18 , Kazuhiro Haginoya 19 , Muzhirah Haniffa 15 , Nozomi Hiraishi 20 , Yoko Hiraki 21 , Satoru Ikemoto 22 , Atsuro Daida 22 , Shin-Ichiro Hamano 22 , Masaki Miura 23, 24 , Akihiko Ishiyama 23 , Osamu Kawano 25 , Akane Kondo 26 , Hiroshi Matsumoto 27 , Nobuhiko Okamoto 28 , Tohru Okanishi 14, 29 , Yukimi Oyoshi 23 , Eri Takeshita 23 , Toshifumi Suzuki 6 , Yoshiyuki Ogawa 30 , Hiroshi Handa 30 , Yayoi Miyazono 31 , Eriko Koshimizu 2 , Atsushi Fujita 2 , Atsushi Takata 2 , Noriko Miyake 2 , Takeshi Mizuguchi 2 , Naomichi Matsumoto 2
Human Mutation ( IF 3.3 ) Pub Date : 2020-11-01 , DOI: 10.1002/humu.24129 Yuri Uchiyama 1, 2 , Daisuke Yamaguchi 3 , Kazuhiro Iwama 2, 4 , Satoko Miyatake 2, 5 , Kohei Hamanaka 2 , Naomi Tsuchida 1, 2 , Hiromi Aoi 2, 6 , Yoshiteru Azuma 2 , Toshiyuki Itai 2 , Ken Saida 2 , Hiromi Fukuda 2, 7 , Futoshi Sekiguchi 2 , Tomohiro Sakaguchi 2 , Ming Lei 2 , Sachiko Ohori 2 , Masamune Sakamoto 2, 4 , Mitsuhiro Kato 8 , Takayoshi Koike 9 , Yukitoshi Takahashi 9 , Koichi Tanda 10 , Yuki Hyodo 11 , Rachel S Honjo 12 , Debora Romeo Bertola 12 , Chong Ae Kim 12 , Masahide Goto 13 , Tetsuya Okazaki 14 , Hiroyuki Yamada 14 , Yoshihiro Maegaki 14 , Hitoshi Osaka 13 , Lock-Hock Ngu 15 , Ch'ng G Siew 15 , Keng W Teik 15 , Manami Akasaka 16 , Hiroshi Doi 7 , Fumiaki Tanaka 7 , Tomohide Goto 17 , Long Guo 18 , Shiro Ikegawa 18 , Kazuhiro Haginoya 19 , Muzhirah Haniffa 15 , Nozomi Hiraishi 20 , Yoko Hiraki 21 , Satoru Ikemoto 22 , Atsuro Daida 22 , Shin-Ichiro Hamano 22 , Masaki Miura 23, 24 , Akihiko Ishiyama 23 , Osamu Kawano 25 , Akane Kondo 26 , Hiroshi Matsumoto 27 , Nobuhiko Okamoto 28 , Tohru Okanishi 14, 29 , Yukimi Oyoshi 23 , Eri Takeshita 23 , Toshifumi Suzuki 6 , Yoshiyuki Ogawa 30 , Hiroshi Handa 30 , Yayoi Miyazono 31 , Eriko Koshimizu 2 , Atsushi Fujita 2 , Atsushi Takata 2 , Noriko Miyake 2 , Takeshi Mizuguchi 2 , Naomichi Matsumoto 2
Affiliation
|
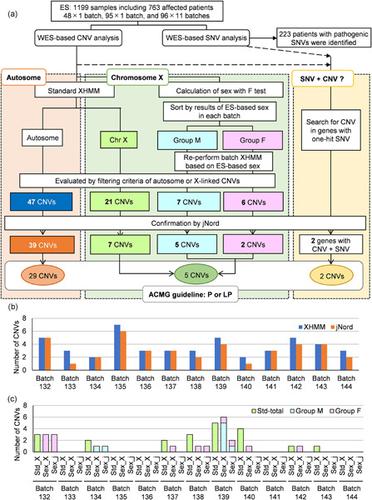
Many algorithms to detect copy number variations (CNVs) using exome sequencing (ES) data have been reported and evaluated on their sensitivity and specificity, reproducibility, and precision. However, operational optimization of such algorithms for a better performance has not been fully addressed. ES of 1199 samples including 763 patients with different disease profiles was performed. ES data were analyzed to detect CNVs by both the eXome Hidden Markov Model (XHMM) and modified Nord's method. To efficiently detect rare CNVs, we aimed to decrease sequencing biases by analyzing, at the same time, the data of all unrelated samples sequenced in the same flow cell as a batch, and to eliminate sex effects of X‐linked CNVs by analyzing female and male sequences separately. We also applied several filtering steps for more efficient CNV selection. The average number of CNVs detected in one sample was <5. This optimization together with targeted CNV analysis by Nord's method identified pathogenic/likely pathogenic CNVs in 34 patients (4.5%, 34/763). In particular, among 142 patients with epilepsy, the current protocol detected clinically relevant CNVs in 19 (13.4%) patients, whereas the previous protocol identified them in only 14 (9.9%) patients. Thus, this batch‐based XHMM analysis efficiently selected rare pathogenic CNVs in genetic diseases.
中文翻译:

使用外显子组数据有效检测拷贝数变异:基于批次和性别的分析
已经报道了许多使用外显子组测序 (ES) 数据检测拷贝数变异 (CNV) 的算法,并评估了它们的灵敏度和特异性、再现性和精确度。然而,尚未完全解决此类算法的操作优化以获得更好的性能。对 1199 个样本进行了 ES,其中包括 763 名具有不同疾病特征的患者。通过 eXome 隐马尔可夫模型 (XHMM) 和改进的 Nord 方法分析 ES 数据以检测 CNV。为了有效检测稀有 CNV,我们旨在通过同时分析在同一流动槽中作为批次测序的所有无关样本的数据来减少测序偏差,并通过分析女性和男性序列分开。我们还应用了几个过滤步骤来更有效地选择 CNV。在一个样本中检测到的 CNV 的平均数量小于 5。这种优化与 Nord 方法的靶向 CNV 分析一起确定了 34 名患者 (4.5%, 34/763) 的致病性/可能致病性 CNV。特别是,在 142 名癫痫患者中,目前的方案在 19 名(13.4%)患者中检测到了临床相关的 CNV,而之前的方案仅在 14 名(9.9%)患者中发现了它们。因此,这种基于批次的 XHMM 分析有效地选择了遗传疾病中罕见的致病性 CNV。而之前的方案仅在 14 名 (9.9%) 患者中发现了它们。因此,这种基于批次的 XHMM 分析有效地选择了遗传疾病中罕见的致病性 CNV。而之前的方案仅在 14 名 (9.9%) 患者中发现了它们。因此,这种基于批次的 XHMM 分析有效地选择了遗传疾病中罕见的致病性 CNV。
更新日期:2020-12-26
中文翻译:

使用外显子组数据有效检测拷贝数变异:基于批次和性别的分析
已经报道了许多使用外显子组测序 (ES) 数据检测拷贝数变异 (CNV) 的算法,并评估了它们的灵敏度和特异性、再现性和精确度。然而,尚未完全解决此类算法的操作优化以获得更好的性能。对 1199 个样本进行了 ES,其中包括 763 名具有不同疾病特征的患者。通过 eXome 隐马尔可夫模型 (XHMM) 和改进的 Nord 方法分析 ES 数据以检测 CNV。为了有效检测稀有 CNV,我们旨在通过同时分析在同一流动槽中作为批次测序的所有无关样本的数据来减少测序偏差,并通过分析女性和男性序列分开。我们还应用了几个过滤步骤来更有效地选择 CNV。在一个样本中检测到的 CNV 的平均数量小于 5。这种优化与 Nord 方法的靶向 CNV 分析一起确定了 34 名患者 (4.5%, 34/763) 的致病性/可能致病性 CNV。特别是,在 142 名癫痫患者中,目前的方案在 19 名(13.4%)患者中检测到了临床相关的 CNV,而之前的方案仅在 14 名(9.9%)患者中发现了它们。因此,这种基于批次的 XHMM 分析有效地选择了遗传疾病中罕见的致病性 CNV。而之前的方案仅在 14 名 (9.9%) 患者中发现了它们。因此,这种基于批次的 XHMM 分析有效地选择了遗传疾病中罕见的致病性 CNV。而之前的方案仅在 14 名 (9.9%) 患者中发现了它们。因此,这种基于批次的 XHMM 分析有效地选择了遗传疾病中罕见的致病性 CNV。









































 京公网安备 11010802027423号
京公网安备 11010802027423号